
Editorial
Austin J Biosens & Bioelectron. 2015;1(2): 1007.
FRET-Based Sensors for Kinase Activity: An Increasing Attractiveness
Corentin Spriet* and Jean-François Bodart
University of Lille, Science and Technology, France
*Corresponding author: Corentin Spriet, University of Lille, Science and Technology, TISBio, UMR 8576, CNRS FR3688, Villeneuve d’Ascq, France.
Received: February 09, 2015; Accepted: February 10, 2015; Published: February 12, 2015
Editorial
FRET-based biosensors for enzymatic activity among other tools
Biosensor is a generic term describing the various analytical devices incorporating a biological sensing element. From their emergence in the 80th, they were mainly either sophisticated laboratory machines or easy to use portable devices [1]. From the 90th, a plethora of new tools, corresponding to the biosensor definition, and aiming at detecting enzymatic activities has emerged. They were structured and developed based upon different purposes, for example, depending upon the will to work in living cells, in lysates from human samples, or to benefit from high sensitivity or selectivity. As an illustration of the wide range of applications and technologies, one can mention the use of (1) in vivo, bioluminescent-based sensors [2,3] or (2) in lysates, functionalized gold nanoparticles [4,5], which could provide highthroughput detection with high sensitivity and selectivity, parameters that are mandatory for clinical diagnostics. From them, genetically encoded FRET-based reporters were gaining an increased interest from the biologist community (Figure 1) especially regarding kinase activity measurements, which we focused on in this editorial.
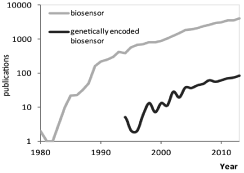
Figure 1: Increasing literature on FRET based biosensors. Comparison of
“biosensor” and “genetically encoded biosensor” searches during the period
1980-2013, using Scopus.
Why such an attractiveness?
These kind of tools overcome the shortcomings of traditional strategies, since enzymatic activities remained mainly assessed in vitro, and characterized in context where spatiotemporal patterns of activities were lost : at best, fixed cells enables snapshots of activity localization, while lysates & fragmented cells only provide average measurement in potentially heterogeneous cells population (Figure 2). Therefore, toolboxes enabling detection of enzymatic activity were welcome in the context of living cells, especially for kinases, which are at the node in molecular networks for many cellular decisions (i.e. cell cycle progression, differentiation, apoptosis).
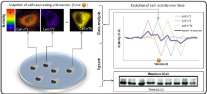
Figure 2: On the different way to look at oscillatory activity. In this scheme,
we represent a population of cells with an oscillatory enzymatic activity,
following the pattern represented in the graph. In this example, we illustrate
the impact of a slight temporal shift between those oscillations. Traditional
biochemical methods rely on lysates of cells population. In such example
the mean activity remains the same and thus, no mean fluctuation of activity
will be measurable. Microscopy based methods allows to measure activity
on the whole field of view or cell by cell. If we measure activity on the whole
field of view, each time point will also give the same mean response but with
a really high standard deviation. To achieve a realistic view of this oscillatory
behavior, one thus needs to perform single cell experiment along time i.e.
microscopy based methods with careful and systematical analysis.
From the basic structure of FRET-based sensor towards KAR
FRET (Förster Resonance Energy Transfer) is a non radiative process involving radiation less energy transfer from a donor fluorophore to an adequately selected and positioned acceptor fluorophore [6]. In the case of genetically encoded FRET-based sensors, the transfer between the donor and the acceptor of the pair can solely occur if they are separated by a distance less than ~10nm. To build such biosensor, both ends are tagged with appropriately chosen fluorophores. Between the fluorophores is located a core made up with peptide consensus site, either targeted by a kinase, a phosphatase, a caspase or ions (calcium for example). Specificity of the FRET-sensor is supported by the specificity of the chosen peptide sequence; Condition is that any modification of the peptide targeted site will alter the biosensor conformation or will cleave it upon either analyte/second messenger presence or protein activity. Thus, a change in FRET efficiency results [7], and can be measured in different manner [8] while it induces modifications of most light properties.
More specifically, in the case of Kinase Activity Reporters (KAR) these tools are composed of the two adapted fluorescent proteins flanking a substrate for a specific kinase and a Phospho-Amino-Acid Binding Domain (PAABD). This PAABD recognizes and binds the phosphorylated substrate, allowing a conformational modification that brings the fluorophores close to each other and leads to a measurable FRET signal. While those sensors are reversible, upon phosphorylation, the PAABD will be released and the FRET signal will decrease accordingly. A docking site may be added to facilitate the interaction between the kinase and its biosensor substrate.
The multiple lives of a KAR
From the discovery of genetically encoded biosensor and its adaptation to numerous fields, a major challenge relies now in the optimization of these tools. Indeed, the first KAR only allowed monitoring massive changes in the analyte’s behavior. First efforts have thus been put through increasing their dynamic range and sublocalizing them in cells compartments. Then, the latest generation sensors allow dissecting low or compartmentalized kinase activity. This optimization step is now part of new biosensors design with the development of dedicated optimization toolkits [9]. Another crucial point is to discriminate fluorescence variation due to the sensor conformational changes from the fluorescence “biological noise and background” [10]. It is thus of crucial importance to make use of “dead” reporters, consisting of the same elements as the reporter to be tested, except for mutations preventing the conformational change due to any changes in the kinase/phosphatase balance. The use of pharmacological activators or inhibitors also allows assess the sensitivity and reversibility of the biosensor, while these chemical tools remain dependent upon their specificity toward kinase(s). These different steps of artifacts controls and intrinsic properties characterization of different KAR version are intended to bring us into gaining more valuable insights regarding kinase activity spatiotemporal profiles, even in micro domains [11].
References
- Turner AP. Biosensors: sense and sensibility. Chem Soc Rev. 2013; 42: 3184-3196.
- Herbst KJ, Allen MD, Zhang J. Luminescent kinase activity biosensors based on a versatile bimolecular switch. J Am Chem Soc. 2011; 133: 5676-5679.
- Van TN, Pellerano M, Lykaso S, Morris MC. Fluorescent protein biosensor for probing CDK/cyclin activity in vitro and in living cells. Chembiochem. 2014; 15: 2298-2305.
- Yin H, Sun B, Dong L, Li B, Zhou Y, Ai S. A signal "on" photoelectrochemical biosensor for assay of protein kinase activity and its inhibitor based on graphite-like carbon nitride, Phos-tag and alkaline phosphatase. Biosens Bioelectron. 2015; 64: 462-468.
- Cui W, Parker LL. A time-resolved luminescence biosensor assay for anaplastic lymphoma kinase (ALK) activity. Chem Commun (Camb). 2015; 51: 362-365.
- Sipieter F, Vandame P, Spriet C, Leray A, Vincent P, Trinel D, et al. From FRET imaging to practical methodology for kinase activity sensing in living cells. Prog Mol Biol Transl Sci. 2013; 113: 145-216.
- Li IT, Pham E, Truong K. Protein biosensors based on the principle of fluorescence resonance energy transfer for monitoring cellular dynamics. Biotechnol Lett. 2006; 28: 1971-1982.
- Newman RH, Zhang J. The design and application of genetically encodable biosensors based on fluorescent proteins. Methods Mol Biol. 2014; 1071: 1-16.
- Fritz RD, Letzelter M, Reimann A, Martin K, Fusco L, Ritsma L, et al. A Versatile Toolkit to Produce Sensitive FRET Biosensors to Visualize Signaling in Time and Space. Science Signaling. 2013; 6.
- Vandame P, Spriet C, Riquet F, Trinel D, Cailliau-Maggio K, Bodart JF. Optimization of ERK activity biosensors for both ratiometric and lifetime FRET measurements. Sensors (Basel). 2014; 14: 1140-1154.
- Vandame P, Spriet C, Trinel D, Gelaude A, Caillau K, Bompard C, et al. The spatio-temporal dynamics of PKA activity profile during mitosis and its correlation to chromosome segregation. Cell Cycle. 2014; 13: 3232-3240.