Special Article - Leukemia
Ann Carcinog. 2017; 2(1): 1008.
Differential Effects of AZD-1208 and SMI-4a, Two Pim-1 Kinase Inhibitors on Primary HAM/TSP and ATL Cells
Jean-Baptiste D1,2, Belrose G1,2, Meniane JC³, Lézin A1,2, Jeannin S4, Mesnard JM5, Olindo S4, Peloponese JM Jr5* and Césaire R1,2*
¹Laboratory of Virology, Martinique University Hospital, France
²EA 4537, Antilles University, France
³Service d’Hématologie Clinique, Martinique University Hospital, France
4Departments of Neurology, Radiology, Vascular Surgery, Martinique University Hospital, France
5IRIM (ex-CPBS)-UMR 9004, Research Institute in Infectiology of Montpellier, University Montpellier, France
*Corresponding author: Raymond Césaire, Laboratoire de Virologie, Centre Hospitalier Universitaire de Fort-de-France, BP 632, 97261 Fort-de-France, Martinique, France
Jean-Marie Péloponèse, Institut de Recherche en Infectiologie de Montpellier (ex CPBS) UMR9004 CNRS, 1919 Route de Mende, 34293 Montpellier Cedex 5, France
Received: February 09, 2017; Accepted: April 18, 2017; Published: April 25, 2017
Abstract
Adult T-cell Leukemia-lymphoma (ATL), an aggressive neoplasm etiologically associated with HTLV-1, is a chemoresistant malignancy. Proviral integration site for Moloney murine leukemia virus-1 (Pim-1) is a critical enzyme that is involved in cell growth, differentiation, survival, apoptosis, senescence and drug resistance. Interaction of Pim-1 with different proteins and association with various signaling pathways make it one of the important antitumor targets. Aberrant elevation of Pim-1 kinase is associated with numerous types of cancer. In this study, we showed that Pim-1 kinase is highly expressed in ATL, as well as in HTLV-1-associated myelopathy/tropical spastic paraparesis (HAM/TSP). Numerous Pim-1 inhibitors are under preclinical studies or clinical trials, such as AZD1208. An increasing number of new Pim-1 inhibitors are still developing and undergoing preclinical investigations. Next, we compared the effect of two PIM-1 inhibitors, AZD-1208 and SMI-4a on HTLV-1 -derived cells lines and ex vivo cultured primary HAM/TSP and ATL leukemic cells. Our results show a differential effects between AZD on survival and proliferation of vs. HTLV-1 derived cells lines. Our results underscore the strong therapeutic potential of Pim kinase inhibition for the treatment of HTLV related pathogenesis such as HAM/TSP and ATL 3
Keywords: PIM-1; Adult T-cell leukemia; HAM/TSP
Introduction
Adult T-cell Leukemia (ATL) is caused by Human T-Lymphotropic Virus-1 (HTLV-1), which is also the etiologic agent of HTLV-1-Associated Myelopathy/Tropical Spastic Paraparesis (HAM/TSP) [1,2]. The estimated lifetime risk of developing ATL in HTLV-1 carriers is 2–7%, and the disease occurs at least 20–30 years after the HTLV-1 infection [3]. ATL is classified as a peripheral T-lymphocytic malignancy of CD4+ T phenotype. The diversity in clinical features and evolution has led to its classification into 4 clinical subtypes: smoldering, chronic, acute, and lymphoma-type ATL [4]. Patients with acute or lymphoma forms have High-Risk ATL (HR-ATL). HR-ATL has a very poor prognosis because of multidrug resistance phenotype of ATL cells, rapid proliferation and large tumor burden, hypercalcemia, and/or infectious complications subsequent to reduced immunologic competence [5,6]. The combination of zidovudine and interferon-alpha with chemotherapy has slightly improved survival in HR-ATL [7,8], but prognosis still remains very poor with three years survival of less than 30% and high relapse rate [9]. In these aggressive forms, allogeneic hematopoietic stem-cell transplantation may improve survival rates [10] and to prevent relapse, Okamura et al. have suggested the possibility of a graft-versus-ATL and graft-versus-HTLV-1 [11].
New therapeutic agents are needed to treat and to improve ATL outcome. Some well-known molecular hallmarks of ATL cells are essential to consider for innovative treatment strategies [12]. The HTLV-1 proviral genome is characterized by the pX region between env and the 3’ Long Terminal Repeat (LTR). The pX-encoded Tax protein activates viral transcription, but is also considered as an oncoprotein [4,13]. Tax has been extensively studied, as a key player at the initial phase of the multistep process of HTLV-1 leukemogenesis. Tax deregulates many cellular signaling pathways related with cell cycle and apoptosis. Tax is pro-mitotic and propels CD4+ T-cell into proliferation [4,13]. At the same time, Tax is the immune-dominant target recognized by the CTL response [14]. Interestingly, tax gene is frequently inactivated in 4 ATL cells [4,13,15]. The HTLV-1 Basic leucine Zipper factor (HBZ), encoded by the pX minus strand is also suspected of down-regulating Tax transcription and contributing to immune escape [16]. HBZ remains the only gene that is consistently expressed in all ATL cases [15,17,18], and is able to induce T-cell lymphoma in transgenic mice [19]. HBZ modulates several cell signaling pathways involved in cell growth and differentiation [9]. HBZ mRNA promotes CD4+ T-lymphocyte proliferation. Evidences are accumulating about the critical role of HBZ in the maintenance of HTLV-1-induced transformation [9]. Proviral integration site for Moloney murine leukemia virus-1 (Pim-1) kinase is observed to interact with numerous proteins participating in various signaling pathways [20-23]. The Pim-1 gene was originally identified as a proviral integration site for Moloney murine leukemia virus-1. Pim- 1 is a proto-oncogene that encodes a serine/threonine kinase, which has a crucial role in oncogenesis [20-24]. This proto-oncogene was originally found in hematopoietic cells as a member of the Pim family (Pim-1, Pim-2 and Pim-3). Transcription of Pim-1 can be activated by several interleukins, such as interleukin-2 (IL-2), IL-3 and IL- 6. It has been shown that the Pim-1 kinase has an essential role in cytokine-induced signal transduction by controlling transcription factors [20,22,24,25] Upregulation of Pim-1 is correlated with cell proliferation induced by mitogens or cytokines, while downregulation of Pim-1 is correlated with growth retention due to the absence of cytokines [20,22,24,25]. Additionally, deficiency of Pim-1 kinase leads to failure in cell survival and growth [20,22,24,25]. Recent studies have shown that Pim-1 is required in drug resistance and has important roles in prostate cancer [26]. Inhibition of Pim kinase activity provides a novel therapeutic approach to the treatment of cancer. In this study, we evaluated the effect of two second generation PIM-1 inhibitor, AZD-1208 and SMI-4a, on HTLV-1 derived cell lines and on primary cells isolated from Asymptomatic Carrier (AC), HAM/TSP and ATL patients. Our finding shows that AZD-1208 preferentially reduces cell proliferation from HAM/TSP patients while SMI-4a 5 inhibits the growth of ATL cells during short-term culture. These results underscore the therapeutic potential of Pim kinase inhibition for the treatment of HTLV related diseases such as HAM/TSP and ATL.
Patients, Materials and Methods
Patients
Blood samples from HTLV-1 infected patients and non-infected donors were obtained from the CHU of Fort-de-France in Martinique. Patients suffering from ATL or HAM/TSP were recruited according to World Health Organization (WHO) criteria. AC had no neurologic or hematological symptoms. According to the French Bioethics laws, the collection of samples from ATL, HAM/TSP and AC has been declared to the French Ministry of Research. Because the protocol is non-interventional (e.g. blood samples collected for routine health care with no additional samplings or specific procedures for subjects), no informed consent was required, as stated by the French Public Health code and the study was conducted anonymously. Clinical collection of samples for research purpose is stored at the Center of Biological Resources of Martinique (CeRBiM).
Cell culture and PIM-1 inhibitors treatment
HTLV-I negative Jurkat, CEM and HTLV-I-positive MT-2, HUT102, C81-66 human T-cell lines, were propagated in RPMI 1640 with 10% fetal calf serum (FCS). PBMC were isolated from EDTAanticoagulated blood samples on Ficoll-density gradients, and washed in phosphate-buffered saline (PBS). CD8+ cells were removed using anti-CD8 paramagnetic microbeads, following the manufacturer’s instructions (Miltenyi Biotec, Paris, France). CD8+-cell–depleted PBMC were then placed in culture wells (round-bottomed 24-well plate) at 106/mL in 1 mL RPMI 1640 medium, supplemented with 10% fetal calf serum, glutamine (2 mmol/L), penicillin (100 IU/mL), and streptomycin (100 μg/mL) (Eurobio, Paris, France). AZD-1208,a benzylidene-1,3-thiazolidine-2,4-dione, and SMI-4a, (5Z)-5-[[3- 7(Trifluoromethyl)phenyl]methylene]-2,4-thiazolidinedione,(Z)- 5-(3-Trifluoromethylbenzylidene) thiazolidine-2,4-dione (Sigma Aldrich) were diluted in dimethyl sulfoxide (Sigma-Aldrich, St. Louis, MO). AZD-1208 and SMI-4a were diluted into the medium at the indicated concentration. Cells and culture supernatants were harvested after different times of incubation at 37°C in 5% CO2, from day 0 (D0) up to D5, depending on the analysis performed.
Flow-cytometry analysis of apoptosis
Cells were washed in PBS, resuspended in annexin V-binding buffer, and incubated for 15 minutes at room temperature with fluorescein–isothiocyanate (FITC)-labeled annexin V (annexin) and propidium iodide (PI) reagents (BD Biosciences, San Jose, CA). 100,000 events in dual-labeled samples were analysed using flow cytometer (FACSCalibur, BD Biosciences). Percentages of viable and apoptotic cells were determined using Cellquest software (Becton-Dickinson Immunocytometry Systems, San Jose, CA) after appropriate compensations.
RNA isolation and qRT-PCR analysis
Cells were collected and cryopreserved as dry pellets until used. Nucleic acid was extracted using the Qiagen AllPrep DNA/RNA Mini Kit (Qiagen, Courtabeouf, France). To obtain first-strand cDNA, total RNA isolated from each sample was subjected to reverse transcription by Superscript II reverse transcriptase (Invitrogen, Cergy Pontoise, France) in the presence of oligo-dT 12-18 primer (Invitrogen). Realtime PCR was run in triplicate using Light Cycler 480 SYBR Green I Master Mix on Light Cycler 480 thermocycler (Roche Applied Science, Meylan, France). Relative mRNA quantification was performed using Cp (crossing point) 8 determined by the 2nd derivative peak of each amplification curve and normalized to housekeeping genes Hypoxanthine-guanine Phosphoribosyltransferase-1 (HPRT1) (forward primer: 5’ TGACACTGGCAAAACAATGCA-3’, reverse primer 5’-GGTCCTTTTCACCAGCAAGCT-3’). qRTPCR. Primer for PIM-1 were purchased from Biorad compagny.
CSFE Proliferation Assays
Cells were stained before culture with 5 μM of 5,6-Carboxyfluorescein diacetate Succinimidyl Ester (CFSE), according to the manufacturer’s instructions (Invitrogen). During 5 days of culture, cells were harvested every 24h, washed twice with PBS, and incubated with Peridinin chlorophyll labeled anti-CD3 and allophycocyanin-labeled anti-CD4. The CFSE fluorescence intensity was measured using FACSCanto II and ModFit LT 4.0 software (Verity Software House, Topsham, ME, USA). Proliferation was evaluated through the proliferation index (i.e. average number of cells that each original cell became), and the non-proliferative fraction (i.e. percent of cells that did not proliferate).
CCK8-Cell proliferation Assay
Cell proliferation was determined using the Cell Counting Kit- 8 (CCK-8) (Dojindo) according to manufacturer’s protocol. Briefly, 104/well cells were plated in 96-well plates. 10 μl WST-8 solutions (Dojindo) were added daily to each well and incubated for 4 hours. The cell viability in each well was determined by reading the OD at 450 nm.
Western blot analysis 9 Whole-cell lysates were prepared using RIPA buffer [10 mM Tris–HCl (pH 7.4), 150 mM NaCl, 1% NP-40, 1 mM EDTA, 0.1% SDS and 1 mM DTT], separated by electrophoresis on SDS-PAGE gels, and transferred onto PVDF membranes (Millipore). Incubations with primary antibodies to detect PIM-1 (Santa Cruz Biotech) and Actine (Sigma-Aldrich) were followed by incubations with the appropriate secondary antibodies conjugated with Horseradish Peroxidase (HRP) (GE Healthcare) and by detection using enhanced luminescence (Roche).
Statistical analysis
(1) Pearson’s correlation for two-dimensional hierarchical clustering analysis; (2) two-tailed pared Student’s t test or 2-way ANOVA for in vitro cell lines and primary cells experiments, including qRT-PCR, cell growth assay. Data are presented as mean ▒ SD. Differences were considered significant at *P<0.05, **P<0.01, and ***P<0.001. 10.
Results
Pim-1L is overexpressed in cells isolated from HAM/TSP and ATL patients
The pim family genes were first identified as proviral integration sites for Moloney murine leukemia virus, but have later been shown to be involved in development of human lymphoid malignancies as well as solid tumors [24]. Aberrant elevation of Pim-1 kinase is associated with numerous types of cancer [24]. We first analyzed expression of PIM-1 in HTLV-1 related cell lines and in primary PBMCs from HTLV-1 infected patients (Figure 1). qRT-PCR analysis reveal that HTLV-1 related cell lines HUT102 and C81-66 expressed significantly (p< 0,0001) more messenger for PIM-1 than unrelated T-cells lines (Figure 1A). Next, we followed by quantitative RTPCR, the expression of pim-1 during culture of CD8+-cell–depleted PBMCs from HTLV-1 carriers without malignancy (AC), HAM/ TSP patients and ATL patients with acute subtype (Figure 1B). In cells derived from AC, we measured low level of pim-1. In contrast in HAM/TSP and in ATL leukemic cells, PIM-1-mRNA expression was spontaneously detectable (Figure 1B). Then, we analyzed PIM1 protein level by western Blot and confirmed that CD8+-depleted PBMCs from HAM/TSP and ATL express high level of PIM-1L in comparison to PBMCs from AC (p<0,0001) and no significant difference in PIM1-L protein level was observed between cells from HAM/TSP and ATL patients (Figure 1C and 1D).
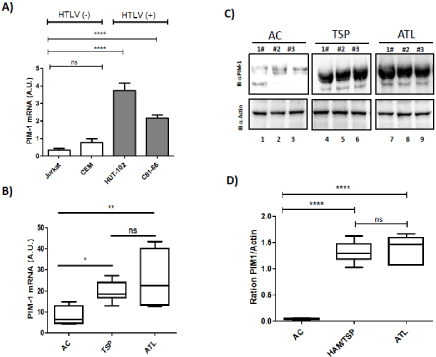
Figure 1: Primary ATL patient samples are characterized by high expression of PIM-1L isoforme. (A) Relative expression of PIM-1 gene was measured by
quantitative RT-PCR and normalized to HPRT RNA levels in control T cell lines (Jurkat, CEM) and HTLV-transformed cell lines (HUT102 and C8166). Significance
difference in relative expression is indicated by asterisk (*p<0.01; *** p<0.0001). (B) CD8+-cell–depleted PBMCs from HTLV-1 Asymptomatic Carriers (AC) and
patients with acute ATL (ATL). Significance difference in relative expression is indicated by asterisk (*p<0.01; *** p<0.0001). (C-D) Western blot analysis of whole cell
extracts prepared from a control cell from asymptomatic carriers (AC), HAM/TSP patients (HAM/TSP) and acute ATL patients (ATL). Two dominant PIM-1-reactive
bands were found, a 39-kDa full-length JunD isoform (PIM-1L) and a shorter 33-kDa isoform (PIM-1S) in HAM/TSP and in ATL. PIM-1L and Actin band intensity
were quantified using Image J software.
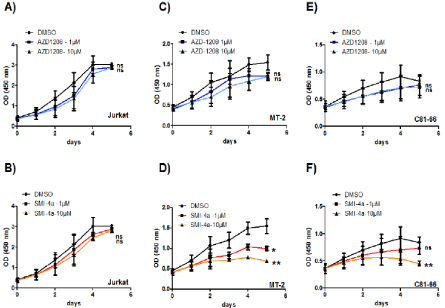
Figure 2: Loss of proliferation in HTLV-derived cell lines treated by the Pim-kinase inhibitors. Cell counts were repeated at least three times. Results represent the
proportion of cells alive after 5 days of AZD-1208 and SMI-4a inhibitor treatment, compared to 5 days treated with DMSO. HTLV-derived cell lines MT-2 (C-D) and
C81-66 (E-F) were treated with 1 or 10 μM of SMI-4a or AZD-1208. Jurkat cells were used as a HTLV-1 negative control (A-B). Significance difference in relative
expression is indicated by asterisk (*p<0.01; *** p<0.0001).
Differential effect of AZD-1208 and SMI-4a on cells isolated from HAM/TSP and ATL patients
The high level of expression of PIM1 and the reported synergistic effect of this kinases with c-MYC in several cancers [24,25], prompted us to test the efficiency PIM-1 inhibition, in preventing proliferation of HTLV-1-infected cells. Numerous Pim-1 inhibitors, such as 11 flavonoid inhibitors ETP-45299 [27], SGI-1776 [28], AZD1208 [29] and SMI-4A [30], have been developed and are now evaluated in preclinical trial [21]. They can be classified as first generation inhibitor (i.e. SGI-1776) and second generation inhibitor (i.e. AZD1208 and SMI4A) [31]. AZD-1208 is a thiazolidene derivative, highly selective, and orally available Pim kinase inhibitor that effectively inhibits all three isoforms of PIM kinase at <5 nM or <150 nM in enzyme and cell assays, respectively. AZD-1208 have been shown to also inhibit the growth of acute myeloid leukemia (AML) cell lines (MV4:11, K562 and U937) [32,33]. SMI-4a, is a benzylidene-thiazolidene-2, 4-dione that inhibits selectively PIM1 and induced G1 arrest in prostate (PC3, DU145) [34] and AML cell lines through inhibition of Cdk2 and translocation of the PIM1 substrate p27kip1 [32,33]. SMI-4A is a novel benzylidene-thiazolidine-2, 4-dione small molecule inhibitor of the Pim kinases, it kills a wide range of both myeloid and lymphoid cell lines with precursor T-cell lymphoblastic leukemia/lymphoma (pre-T-LBL/T-ALL) being highly sensitive [28]. To test the effect of those two second generation inhibitors, one control T-cell line Jurkat and two HTLV-1-transformed T-cells MT-2 and C81-66 were treated with either AZD-1208, or SMI-4a (1 to 10 μM) and cell viability was assessed over a period of five days (Figure 2). Interestingly, Jurkat cells were mostly resistant to the two drugs (Figure 2A and 2B). Compared with control treatment (DMSO) or to inhibitor-treated control T-cells, proliferation of HTLV-1-transformed cells decreased more significantly when treated with SMI-4a (Figure 2D and 2F) than with AZD-1208 (Figure 2C and 2E). This finding indicates that PIM-1 function may be selectively important for the survival of these HTLV- 1-transformed T-cells as compared with control T-cells and that SMI- 4a could be an interesting new therapeutic agent to treat ATL.
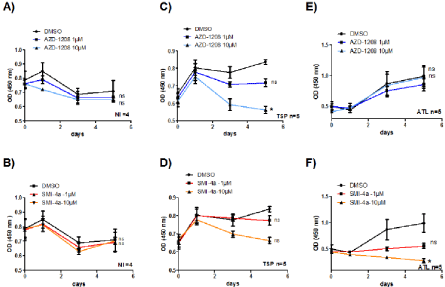
Figure 3: PIM-1 inhibitor SMI-4a but not AZD-1208 abolished ATL-cell growth. CD8+-depleted PBMCs isolated from four non-infected individuals (A-B) or five
HAM/TSP patients (C-D) and five ATL patients (E-F) were treated as indicated with either AZD-1208 (1-10 μM) or SMI-4a (1-10 μM) for 5 days. Cell viability was
measured using WST-8 (2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium, monosodium salt) coloration (Dojindo) according to
the manufacturer’s protocol. Significance difference in relative expression is indicated by asterisk (*p<0.01; *** p<0.0001).
We next checked to see if PIM-1 also plays an important proliferative role in CD8+-depleted PBMCs isolated from HTLV- 1-infected patients. AC, HAM/TSP and ATL leukemic cells were treated with either AZD-1208, or SMI-4a (1 to 10 μM) and the proliferation of 12 inhibitor-treated cells was followed over a period of 5 days (Figure 3). Compared with control treatment (DMSO) or to inhibitor-treated control T-cells, proliferation of HAM/TSP cells decreased more significantly when treated with AZD-1208 than with SMI-4a (Figure 3C and 3D) while ATL cells were more sensitive to apoptosis when treated with SMI-4a than AZD-1208 (Figure 3E and 3F).

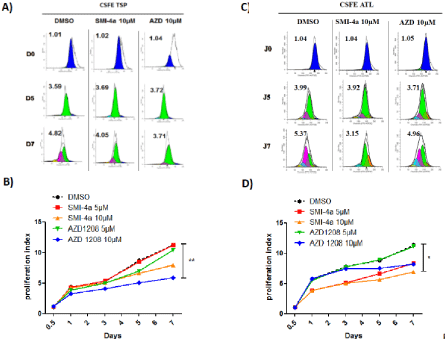
Figure 4: PIM-1 inhibitor SMI-4a but not AZD-1208 abolished ATL-cell proliferation.
CD8+-depleted PBMCs isolated from HAM/TSP patients (A-B) and ATL patients (C-D) were treated as indicated with either AZD-1208 (5-10 μM) or SMI-4a (510
μM) for 5 days. Cell proliferation was asses using by flow cytometry using CarboxyFluorescein diacetate, Succinimidyl Ester or CFSE staining according to the
manufacturer’s protocol. Significance difference in relative expression is indicated by asterisk (*p<0.01; ***p<0.0001).
To confirm the effect of AZD-1208 and SMI-4a on cellproliferation, we stained CD8+-cell–depleted PBMCs from HAM/ TSP and ATL patients with CFSE, followed by flow cytometry analysis for 7 days. Figure 4A and 4C shows a representative experiment with ModFit histograms obtained after light scatter gating and CD3+/ CD4+ gating to select HTLV-1 infected T-cells. Compared with control treatment (DMSO), proliferation index of HAM/TSP CD4+- T-cells were lower when cells were treated with AZD-1208 than with SMI-4a (Figure 4B), while proliferation of ATL CD4+- T-cells decreased more significantly when treated with SMI-4a than with AZD-1208 (Figure 4D).
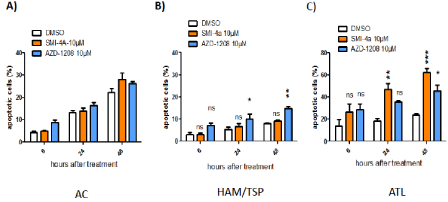
Figure 5: PIM-1 inhibitor SMI-4a induces significant apoptosis in fresh CD8+-depleted PBMCs from ATL patients. SMI-4a induces apoptosis in ATL cells with
minimal cell death in normal PBMCs. AC (A), HAM/TSP (B) or ATL (C) cells were treated with either DMSO; SMI-4a (10 μM),AZD-1208 (10 μM). Apoptosis was
determined by fluorescence activated cell sorting analyses of cells after Annexin V staining.
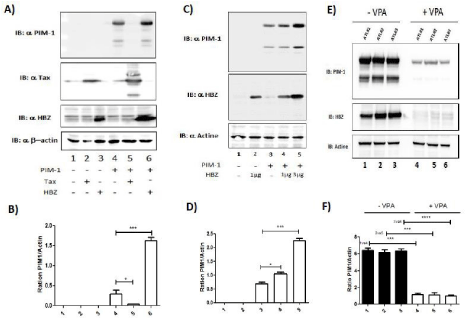
Figure 6: HBZ induces the expression of PIM-1L. (A-B) 293T were co-transfected with PIM-1 plasmid and Tax (lanes 3 & 5) or HBZ plasmid (lanes 4 & 6). Fortyeight
hours after 20 transfection, PIM-1, Tax, HBZ and actin expression were assessed by western blot two dominant PIM-1-reactive bands were found, a 39-kDa
full-length PIM isoform (PIM-1LFL) and a shorter 33-kDa isoform (PIM-1S). PIM-1L and Actin band intensity were quantified using Image J software. Significance
difference in relative expression is indicated by asterisk (*p<0.01; *** p<0.0001). (C-D) 293T were co-transfected with JunD-Flag plasmid and increasing amount
of HBZ-Myc plasmid (lanes 2-4). Forty-eight hours after transfection, PIM-1, HBZ and actin expression were assessed by western blot. PIM-1L and Actin band
intensity were quantified using Image J software. Significance difference in relative expression is indicated by asterisk (*p<0.01; *** p<0.0001). (E-F) Valproate
treatment of CD4+ T cells isolated from ATL patients decrease PIM-1L expression. CD8+-cell–depleted PBMCs from patients with acute ATL (ATL) treated or not
with VPA for 5 days. PIM-1, HBZ and actin expression were assessed by western blot. PIM-1L and Actin band intensity were quantified using Image J software.
Significance difference in relative expression is indicated by asterisk (*p<0.01; *** p<0.0001).
Independently, we also assessed inhibitors-treated cells for apoptosis (Figure 5). Consistent with the proliferation results (Figures 3 and 4), AZD-1208-treated HAM/TSP CD4+- T-cells were significantly increased in Annexin V-positive cells. For example, whereas 7 % of AZD-1208-treated AC cells showed early apoptosis (Annexin V-positive/ PI-negative), the same treatment provoked 15 % of HAM/TSP cells, respectively, into incipient apoptosis (Figure 5B). SMI-4a-treated ATL CD4+- T-cells shown a 40% increase in apoptosis 48h after treatment (Figure 5C). This finding indicates that PIM-1 function may be selectively important for the survival of HTLV-1-infected primary T-cells as compared with control T-cells or AC. Together our results underscore the therapeutic potential of Pim-1 kinase inhibition for the treatment of acute ATL.
Opposite effects of Tax and HBZ on Pim-1 expression 13
In order to understand the difference of sensitivity to Pim-1 inhibitors between HAM/TSP and ATL cells, we measured the effect of the two viral proteins Tax and HBZ on the expression of PIM- 1. To mimic the three different population of HTLV -infected cells, HEK293T were cotransfected with PIM-1 alone (Figure 6A lane 4) or with Tax (Figure 6A lane 5) or with HBZ (Figure 6A lane 6). Interestingly, expression level of PIM-1 was drastically reduced by Tax (Figure 6A and 6B lane 5). On the opposite, HBZ induced in a dose dependent manner expression of PIM-1 protein (Figure 6C and 6D).
We next tested how extinction of HBZ expression would impact PIM-1 expression. As we previously described, treatment with Valproic Acid (VPA), a histone deacetylase inhibitor, impaired HBZ expression in primary HTLV infected cells [35]. CD8+ depleted PBMCs from ATL were cultured with 1mM VPA for 5 days. Using WB, we measured the expression levels of PIM-1 and HBZ in CD8+ depleted PBMCs from acute ATL patients (Figure 6E). We found that VPA treatment resulted in a significant decrease of PIM-1L expression (Figure 6F) concomitant with the loss of HBZ expression. These observations suggest that by enhancing PIM-1L expression, HBZ might play a role in the observed chemoresistance of ATL cells.
Discussion and Conclusion
ATL is an incurable and poorly treatable disease. Despite advances in both chemotherapy and supportive care, median survival time of patients remains less than 1 year. As pointed out by Yamada and Tomonaga, an important amount knowledge in molecular biology and oncogenesis of ATL has accumulated but has not yet been translated into improved prognosis of affected patients [36]. In fact, it has been reported that the prognosis of indolent subtypes, chronic and smoldering ATL, was 4.1 years, which is poorer than 14 previously thought [37]. New therapeutic approaches are needed to treat and to improve ATL outcome.
In this study, we investigated the ability of two second generation PIM-1 inhibitors SMI-4a and AZD-1208 to kill HTLV-1-infected cells. We find that all tested cells are sensitive to these agents with some differences in regards of the HTLV-induced pathology. Indeed, we observed that cells acutely infected by HTLV-1 (i.e. MT-2 and CD8+ depleted PBMCs from HAM/TSP patients) were sensitive to AZD-1208 treatment while HTLV-1 chronically infected cells (i.e. C81-66 and CD8+ depleted PBMCs from ATL patients) were more sensitive to SMI-4a (Figures 2-5). Interestingly, we also observed that the two viral proteins Tax and HBZ have different effect on PIM-1L expression. Indeed the observed degradation of PIM-1L induced by Tax which is mainly express in MT-2 and CD8+ depleted PBMCs from HAM/TSP might explain why those cells are not sensitive to SMI-4a. Indeed while AZD-1208 is targeting all three isoformes of PIM Kinase, SMI-4a is highly specific to PIM-1L [30]. Emerging evidence has shown that Pim-1L kinase has been associated with the drug-resistant abilities of cancer cells [25]. Pim-1L mediates drug resistance through interaction with and phosphorylation of Etk, P-glycoprotein (Pgp), Breast Cancer Resistant Protein (BCRP) [20]. The original findings on Pim-1-mediated drug resistance come from the early study that Pim-1 overexpression allows cells to undergo prolonged survival upon withdrawal of IL-3 [22]. Following this, Pim-1L-mediated drug resistance was identified as a mechanism of inhibiting p53-induced apoptosis [20]. Mechanistically, Pim-1L competes with p53 to bind non-receptor tyrosine kinase Etk. Etk signaling has an important role in this drug resistance as Pim-1L, but not Pim-1S, directly interacts with Etk at the plasma membrane while Etk signaling can promote cell survival by inhibiting p53 [20]. Thus, Pim-1L showed a higher ability to protect cancer cells to undergo apoptosis induced by chemotherapy drugs. Interestingly, while this study was ongoing, Bellon, et al. showed that preclinical testing of 15 AZD-1208 in a mouse model of ATL resulted in significant prevention of ED40515 cells growth in vivo [38]. Discrepancy on the observed effects of AZD-1208 and SMI-4a between our study and Bellon and al. [38] results comes from the use of different HTLV- 1-derived cell lines and by the fact that we used for our cytoxocity assays fresh CD8+-depleted PBMCs isolated from HTLV-1 infected patients and not HTLV- transformed cells lines which might have derived since they were establish in the early 80’s. Nevertheless more preclinical studies of SMI-4a and others targeted at the Pim kinases will be needed to determine whether this protein kinase will be a novel target for therapeutic treatment of ATL and HAM/TSP.
References
- Gessain A, Cassar O. Epidemiological Aspects and World Distribution of HTLV-1 Infection. Front Microbiol. 2012; 3: 388.
- Hinuma Y, Nagata K, Hanaoka M, Nakai M, Matsumoto T, Kinoshita KI, et al. Adult T-cell leukemia: antigen in an ATL cell line and detection of antibodies to the antigen in human sera. Proc Natl Acad Sci USA. 1981; 78: 6476-6480.
- Iwanaga M, Watanabe T, Utsunomiya A, Okayama A, Uchimaru K, Koh KR, et al. Human T-cell Leukemia Virus type I (HTLV-1) proviral load and disease progression in asymptomatic HTLV-1 carriers: a nationwide prospective study in Japan. Blood. 2010; 116: 1211-1219.
- Matsuoka M, Jeang KT. Human T-Cell Leukaemia Virus type 1 (HTLV-1) infectivity and cellular transformation. Nat Rev Cancer. 2007; 7: 270-280.
- Tsukasaki K, Tobinai K. Clinical Trials and Treatment of ATL. Leuk Res Treatment. 2012; 2012: 101754.
- Bazarbachi A, Suarez F, Fields P, Hermine O. How I treat adult T-cell leukemia/lymphoma. Blood. 2011; 118: 1736-1745.
- Bazarbachi A, Ghez D, Lepelletier Y, Nasr R, de The H, El-Sabban ME, et al. New therapeutic approaches for adult T-cell leukaemia. The Lancet Oncology. 2004; 5: 664-672.
- Hodson A, Crichton S, Montoto S, Mir N, Matutes E, Cwynarski K, et al. Use of zidovudine and interferon alfa with chemotherapy improves survival in both acute and lymphoma subtypes of adult T-cell leukemia/lymphoma. J Clin Oncol. 2011; 29: 4696-46701.
- Mesnard JM, Barbeau B, Cesaire R, Peloponese JM. Roles of HTLV-1 Basic Zip Factor (HBZ) in Viral Chronicity and Leukemic Transformation. Potential New Therapeutic Approaches to Prevent and Treat HTLV-1-Related Diseases. Viruses. 2015; 7: 6490-6505.
- Hishizawa M, Kanda J, Utsunomiya A, Taniguchi S, Eto T, Moriuchi Y, et al. Transplantation of allogeneic hematopoietic stem cells for adult T-cell leukemia: a nationwide retrospective study. Blood. 2010; 116: 1369-1376.
- Okamura J, Uike N, Utsunomiya A, Tanosaki R. Allogeneic stem cell transplantation for adult T-cell leukemia/lymphoma. Int J Hematol. 2007; 86: 118-125.
- Yamagishi M, Watanabe T. Molecular hallmarks of adult T cell leukemia. Front Microbiol. 2012; 3: 334.
- Peloponese JM, Kinjo T, Jeang KT. Human T-cell leukemia virus type 1 Tax and cellular transformation. Int J Hematol. 2007; 86: 101-106.
- Goon PK, Biancardi A, Fast N, Igakura T, Hanon E, Mosley AJ, et al. Human T cell Lymphotropic Virus (HTLV) type-1-specific CD8+ T cells: frequency and immunodominance hierarchy. J Infect Dis. 2004; 189: 2294-2298.
- Kataoka K, Nagata Y, Kitanaka A, Shiraishi Y, Shimamura T, Yasunaga J, et al. Integrated molecular analysis of adult T cell leukemia/lymphoma. Nature genetics. 2015; 47: 1304-1315.
- Gaudray G, Gachon F, Basbous J, Biard-Piechaczyk M, Devaux C, Mesnard JM. The complementary strand of the human T-cell leukemia virus type 1 RNA genome encodes a bZIP transcription factor that down-regulates viral transcription. J Virol. 2002; 76: 12813-12822.
- Vicente C, Cools J. The genomic landscape of adult T cell leukemia/lymphoma. Nature genetics. 2015; 47: 1226-1227.
- Satou Y, Yasunaga J, Yoshida M, Matsuoka M. HTLV-I basic leucine zipper factor gene mRNA supports proliferation of adult T cell leukemia cells. Proc Natl Acad Sci USA. 2006; 103: 720-725.
- Satou Y, Yasunaga J, Zhao T, Yoshida M, Miyazato P, Takai K, et al. HTLV-1 bZIP factor induces T-cell lymphoma and systemic inflammation in vivo. PLoS Pathog. 2011; 7: e1001274.
- Isaac M, Siu A, Jongstra J. The oncogenic PIM kinase family regulates drug resistance through multiple mechanisms. Drug resistance updates: reviews and commentaries in antimicrobial and anticancer chemotherapy. 2011; 14: 203-211.
- Blanco-Aparicio C, Carnero A. Pim kinases in cancer: diagnostic, prognostic and treatment opportunities. Biochem Pharmacol. 2013; 85: 629-643.
- Warfel NA, Kraft AS. PIM kinase (and Akt) biology and signaling in tumors. Pharmacology & therapeutics. 2015; 151: 41-49.
- Gharwan H, Groninger H. Kinase inhibitors and monoclonal antibodies in oncology: clinical implications. Nature reviews Clinical oncology. 2016; 13: 209-227.
- Narlik-Grassow M, Blanco-Aparicio C, Carnero A. The PIM family of serine/threonine kinases in cancer. Med Res Rev. 2014; 34: 136-159.
- Tursynbay Y, Zhang J, Li Z, Tokay T, Zhumadilov Z, Wu D, et al. Pim-1 kinase as cancer drug target: An update. Biomedical reports. 2016; 4: 140-146.
- Xie Y, Bayakhmetov S. PIM1 kinase as a promise of targeted therapy in prostate cancer stem cells. Molecular and clinical oncology. 2016; 4: 13-17.
- Blanco-Aparicio C, Collazo AM, Oyarzabal J, Leal JF, Albaran MI, Lima FR, et al. Pim 1 kinase inhibitor ETP-45299 suppresses cellular proliferation and synergizes with PI3K inhibition. Cancer Lett. 2011; 300: 145-153.
- Lin YW, Beharry ZM, Hill EG, Song JH, Wang W, Xia Z, et al. A small molecule inhibitor of Pim protein kinases blocks the growth of precursor T-cell lymphoblastic leukemia/lymphoma. Blood. 2010; 115: 824-833.
- Keeton E, McEachern K, Alimzhanov M, Wang S, Cao Y, Bao L, et al. Abstract 2796: Efficacy and biomarker modulation by AZD1208, a novel, potent and selective pan-Pim kinase inhibitor, in models of acute myeloid leukemia. Cancer research. 2012; 72: 2796.
- Xia Z, Knaak C, Ma J, Beharry ZM, McInnes C, Wang W, et al. Synthesis and evaluation of novel inhibitors of Pim-1 and Pim-2 protein kinases. J Med Chem. 2009; 52: 74-86.
- Mondello P, Cuzzocrea S, Mian M. Pim kinases in hematological malignancies: where are we now and where are we going? Journal of hematology & oncology. 2014; 7: 95.
- Keeton EK, McEachern K, Dillman KS, Palakurthi S, Cao Y, Grondine MR, et al. AZD1208, a potent and selective pan-Pim kinase inhibitor, demonstrates efficacy in preclinical models of acute myeloid leukemia. Blood. 2014; 123: 905-13.
- Chen LS, Yang JY, Liang H, Cortes JE, Gandhi V. Protein profiling identifies mTOR pathway modulation and cytostatic effects of Pim kinase inhibitor, AZD1208, in acute myeloid leukemia. Leuk Lymphoma. 2016; 57: 2863-2873.
- Cervantes-Gomez F, Lavergne B, Keating MJ, Wierda WG, Gandhi V. Combination of Pim kinase inhibitors and Bcl-2 antagonists in chronic lymphocytic leukemia cells. Leuk Lymphoma. 2015; 28: 1-9.
- Belrose G, Gross A, Olindo S, Lezin A, Dueymes M, Komla-Soukha I, et al. Effects of valproate on Tax and HBZ expression in HTLV-1 and HAM/TSP T lymphocytes. Blood. 2011; 118: 2483-2491.
- Yamada Y, Tomonaga M. The current status of therapy for adult T-cell leukaemia-lymphoma in Japan. Leuk Lymphoma. 2003; 44: 611-618.
- Kawano N, Yoshida S, Kuriyama T, Tahara Y, Yamashita K, Nagahiro Y, et al. Clinical Features and Treatment Outcomes of 81 Patients with Aggressive Type Adult T-cell Leukemia-lymphoma at a Single Institution over a 7-year Period (2006-2012). Intern Med. 2015; 54: 1489-1498.
- Bellon M, Lu L, Nicot C. Constitutive activation of Pim1 kinase is a therapeutic target for adult T-cell leukemia. Blood. 2016; 127: 2439-2450.