
Research Article
Austin Diabetes Res. 2024; 9(1): 1032.
Evaluating Machine Learning Models for Early Diabetes Prediction: A Comparative Study
Qazi Waqas Khan*
Department of Computer Engineering, Jeju National University, Jejusi 63243, Jeju Special Self-Governing Province, Republic of Korea
*Corresponding author: Qazi Waqas Khan, Department of Computer Engineering, Jeju National University, Jejusi 63243, Jeju Special Self-Governing Province, Republic of Korea. Email: waqasqazi19@stu.jejunu.ac.kr
Received: July 01, 2024 Accepted: July 18, 2024 Published: July 25, 2024
Abstract
High blood glucose levels can affect the body’s organs, causing blindness, renal illness, and heart and kidney diseases. Globally, Diabetic patients experience a mortality rate of 38% yearly. Machine Learning methods are used in the literature to predict diabetes. The prediction of machine learning models can assist doctors in making early decisions. This study employed the Neural Oblivious Decision Ensembles (NODE), Xtreme Gradient Boosting (XGB), AdaBoost, and Support Vector Machine (SVM) models to diagnose diabetes. An early-risk diabetes dataset is utilized in this study to conduct the experiments. The principal component analysis method is employed to extract the features. The performance metrics for evaluating machine learning classifiers are accuracy, precision, recall, and f score. The experimental results of the learning models show that the XGB model has achieved higher prediction results than the SVM, AdaBoost, and NODE. These findings conclude that the utilization of this approach assists the stakeholders in the diagnosis of early diabetes.
Introduction
High blood sugar is a leading factor of death, depicting diabetes as a destructive chronic illness and creating an alarming condition. According to WHO, the number of diabetic patients increased significantly from 108 million in 1980 to 422 million in 2014 [1]. About 8.5% of adults and 30.3% of the U.S. population are affected by diabetes [2]. China and India, being the most populous countries, have the highest diabetes rates of 98 million and 65.1 million cases, respectively [3]. Both types of diabetes are serious conditions. Type 1 diabetes attacks the pancreas and affects the formulation of insulin in the body; type 2 diabetes includes insulin resistance, which stops the body from using insulin, causing high blood glucose levels [4]. Diabetes cannot be cured but it can be treated, early diagnosis can minimize the complication risks [5]. A balanced diet and early detection can increase an individual’s lifespan. Detecting diabetes at an early stage based on a doctor’s assessment can be inaccurate because of gaps in understanding the related patterns [6]. However, predictive analytics can improve the identification of at-risk individuals, anticipate issues, and enhance treatment results [7]. Predictive analytics can identify high-risk individuals, predict complications, and enhance care. A doctor can determine the most effective treatment course for everyone affected by diabetes, leading to better outcomes [8]. Therefore, a Computer-Aided Diagnosis (CAD) system can help physicians make better decisions for diagnosing diabetes at an early stage. [9]. The CAD system analyzes blood sugar levels, haemoglobin A1C levels, and other useful clinical data to detect diabetes and suggest necessary actions depending on the information obtained.
The Neural Oblivious Decision Ensembles (NODE), Xtreme Gradient Boosting (XGB), AdaBoost, and Support Vector Machine (SVM) models are used in this study to predict diabetes—label encoder method to convert the text category into numeric. The standard scalar method converts the feature value into the same between 0 and 1.
The structure of the paper is described as Section 2 explains the details of a proposed method for diabetes prediction. Sections 3 and 4 explain the experimental results and conclusion of the paper.
Proposed Methods
This section briefly describes the proposed machine learning models used for diabetes prediction. Figure 1 shows the architecture diagram, which shows that we used the early diabetes risk dataset as an input for the data pre-processing module. In data pre-processing, the label encoding, and standard scalar method are applied for data preparation. The prepared data is transmitted as input to proposed models for diabetes classification. The performance of a machine learning model is analyzed using the accuracy, precision, recall, and f score.

Figure 1: Comparison graph of Machine Learning Model for Diabetes Classification (with PCA feature extraction) based on accuracy.
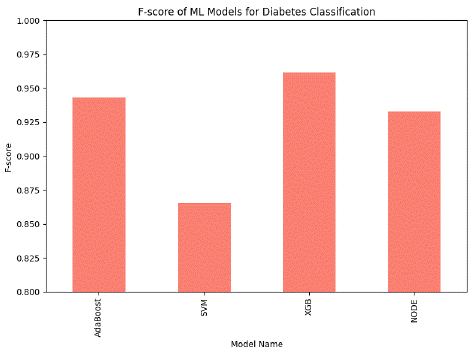
Figure 2: Comparison graph of Machine Learning Model for Diabetes Classification (with PCA feature extraction) based f score.
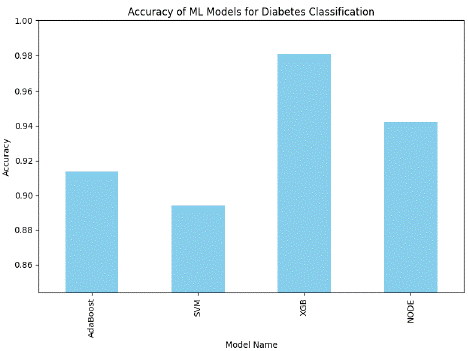
Figure 3: Comparison graph of Machine Learning Model for Diabetes Classification (without PCA feature extraction) based on accuracy.
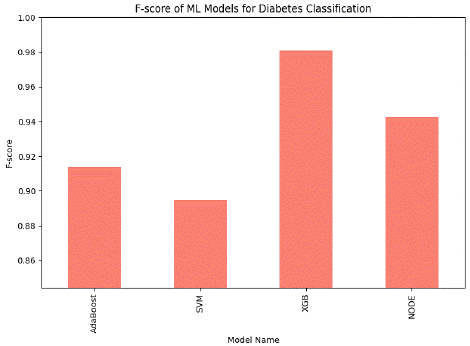
Figure 4: Comparison graph of Machine Learning Model for Diabetes Classification (without PCA feature extraction) based on fs core.
Data Preprocessing
The data preprocessing method cleans and prepares the raw data for the learning model. In this study, the scikit label encoder method converts the text category into numeric, and the z-score method normalizes the data into the same scale.
Machine Learning Models
This study utilized the NODE, XGB, Adaboost, and SVM models to classify diabetes. These models are used with and without the Principal Component Analysis (PCA). The PCA method extracts features and transforms them into n components, where n is a target feature space.
Node Neural Oblivious Decision Ensembles
The NODE [11] is a robust machine-learning architecture that merges decision trees with neural network architectures to improve predictive accuracy. By utilizing an oblivious decision tree, NODE efficiently captures complex patterns in the data, making it highly suitable for classification and regression tasks.
Xtreme Gradient Boosting
Xtreme Gradient Boosting (XGB) [12] is a powerful and effective gradient-boosting framework that outperforms classification and regression. It employs parallel processing and advanced regularization techniques to boost model robustness and prevent overfitting.
AdaBoost
Adaptive boosting, known as AdaBoost [13], is a robust machine-learning algorithm that integrates multiple weak classifiers to create a robust classifier. In a later iteration, it adjusts the weights of misclassified instances, focusing more on challenging cases to enhance the performance for both classification and regression.
Support Vector Machine
The SVM [14] is a supervised machine-learning algorithm that classifies cases by finding the optimum hyperplane that maximizes the margin between data points. It uses the kernel function to map the data into a higher-dimensional feature space for better class separation.
Evaluation Metrics
This study validated the performance of a machine learning model using accuracy, precision, recall, and f-score evaluation metrics.
Results and Discussion
The table presents the experimental outcomes of the proposed classifiers for classifying diabetes with PCA feature extraction. It shows four different classifiers evaluated using the given metrics: Accuracy, Precision, Recall, and F-score. Results explain that XGB is highly effective, surpassing all the models with the highest prediction performance. AdaBoost and NODE also performed well, with high scores across all metrics, and SVM showed lower performance than all other proposed models, with moderate scores across all metrics.
Model Name
Accuracy
Precision
Recall
F score
AdaBoost
0.9423
0.9469
0.9423
0.9431
SVM
0.8654
0.8654
0.8654
0.8654
XGB
0.9615
0.9629
0.9615
0.9618
NODE
0.9327
0.9334
0.9327
0.9330
Table 1: Experimental Results of a Machine Learning Model for Diabetes Classification (with PCA feature extraction).
Model Name
Accuracy
Precision
Recall
F score
AdaBoost
0.9135
0.9143
0.9135
0.9138
SVM
0.8942
0.8952
0.8942
0.8946
XGB
0.9808
0.9819
0.9808
0.9809
NODE
0.9423
0.9439
0.9423
0.9427
Table 2: Experimental Results of a Machine Learning Model for Diabetes Classification (without PCA feature extraction).
The table presents the experimental outcomes of the proposed classifiers for classifying diabetes with PCA feature extraction. It shows four different classifiers evaluated using the given metrics: Accuracy, Precision, Recall, and F-score. Results explain that XGB is highly effective, surpassing all the models with the highest prediction performance. AdaBoost and NODE also performed well, with high scores across all metrics, and SVM showed lower performance than all other proposed models, with moderate scores across all metrics.
This figure compares the F-score of proposed machine-learning models used for diabetes classification with PCA feature extraction. The chart shows that XGB achieves the highest f-score of 96.18% with AdaBoost and NODE having slightly less f-score, while SVM has the lowest f-score among all the proposed classifiers.
This table represents the experimental outcomes of four proposed machine-learning classifiers for classifying diabetes without PCA feature extraction. It can be seen from experimental results that the XGB is a highly effective classifier in this case as well, as it receives the highest score across all the metrics; NODE and AdaBoost also performed well, while SVM shows the lowest performance than other classifiers. The results also indicate that XGB outperforms with or without PCA extraction, SVM and NODE enhance their performance without PCA extraction, and AdaBoost slightly decreases its performance when PCA is not used.
This figure compares the Accuracy of proposed machine-learning models used for diabetes classification without PCA feature extraction. The chart shows that XGB achieves the highest accuracy of 98.08%, demonstrating its effectiveness in diabetes classification; NODE also performs well but is slightly lower than XGB. AdaBoost has a moderate performance, and SVM has the lowest performance rate of all the classifiers.
This figure compares the F-score of proposed machine-learning models for diabetes classification without PCA feature extraction. The chart shows that XGB leads in performance by achieving the highest F-score of 98.09%, showing efficiency in diabetes classification. NODE also performs well but is slightly lower than XGB. AdaBoost has a moderate performance, and SVM has the lowest performance rate of all the classifiers.
Conclusion
The computer-aided diagnosis systems are crucial to diagnose the diseases. These computer-aided diagnosis systems assist doctors in detecting or diagnosing the disease. Diabetes is a chronic disease, and detection of this disease at the earliest stage is crucial to saving a patient's life. This study employed machine learning-based methods for diagnosing diabetes. The experiment shows that the XGB model has achieved higher prediction results than other models. In contrast, the SVM model has achieved lower prediction accuracy than the other proposed models. The experimental findings show that this method helps stakeholders in the early diagnosis of diabetes. In the future, we can utilize some wrapper-based feature selection methods to select the optimal feature from the dataset. Further, we will use the ensemble model for the prediction of diabetes.
References
- Liu J, Ren ZH, Qiang H, Wu J, Shen M, Zhang L, et al. Trends in the incidence of diabetes mellitus: results from the Global Burden of Disease Study 2017 and implications for diabetes mellitus prevention. BMC public health. 2020; 20: 1415.
- Siva C. Factors Affecting Diabetes Rates in The United States. MS thesis. Lamar University-Beaumont. 2019.
- Pradeepa R, Mohan V. Prevalence of type 2 diabetes and its complications in India and economic costs to the nation. European journal of clinical nutrition. 2017; 71: 816-824.
- Eizirik Décio L, Lorenzo Pasquali, Miriam Cnop. Pancreatic β-cells in type 1 and type 2 diabetes mellitus: different pathways to failure. Nature Reviews Endocrinology. 2020; 16: 349-362.
- Saruar A, Hasan K, Neaz S, Hussain N, Hossain F, Rahman T. Diabetes Mellitus: insights from epidemiology, biochemistry, risk factors, diagnosis, complications and comprehensive management. Diabetology. 2021; 2: 36-50.
- Ekaterina J, Spohrer K, Heinzl A, Gawlitza J. Augmenting medical diagnosis decisions? An investigation into physicians’ decision-making process with artificial intelligence. Information Systems Research. 2021; 32: 713-735.
- Zheng Le, Wang O, Hao S, Ye C, Liu M, Xia M, et al. Development of an early-warning system for high-risk patients for suicide attempt using deep learning and electronic health records. Translational psychiatry. 2020; 10: 72.
- American Diabetes Association. 5. Facilitating behavior change and well-being to improve health outcomes: Standards of Medical Care in Diabetes—2020. Diabetes care. 2020; 43: S48-S65.
- Hiroshi F. AI-based computer-aided diagnosis (AI-CAD): the latest review to read first. Radiological physics and technology. 2020; 13: 6-19.
- Usharani T, Umapathy S, Janardhanan K, Thirunavukkarasu R. A computer aided diagnostic method for the evaluation of type II diabetes mellitus in facial thermograms. Physical and Engineering Sciences in Medicine. 2020; 43: 871-888.
- Fazila M, Khan QW, Rizwan A, Alnashwan R, Atteia G. A Machine Learning-Based Framework with Enhanced Feature Selection and Resampling for Improved Intrusion Detection. Mathematics. 2024; 12: 1799.
- Ugochukwu O, Ukwandu E. Machine learning for an explainable cost prediction of medical insurance. Machine Learning with Applications. 2024; 15: 100516.
- Sensen W, Liu W, Yang S, Huang H. An optimized AdaBoost algorithm with atherosclerosis diagnostic applications: adaptive weight-adjustable boosting. The Journal of Supercomputing. 2024; 80: 1-30.
- Khan, QW, Kim BW, Ahmed R, Rizwan A, Khan AN, Kim K, et al. Predictive modeling of water table depth, drilling duration, and soil layer classification using adaptive ensemble learning for cost-effective percussion water borehole drilling. IEEE Access. 2023; 99: 1.