
Research Article
Gastrointest Cancer Res Ther. 2016; 1(1): 1005.
Gene Expression Analysis of Sporadic Early-Onset Rectal Adenocarcinoma
Nfonsam V1,2*, Xu W³, Koblinski J² and Jandova J1,2
¹UA Cancer Center, 1515 N Campbell Avenue, Tucson, AZ 85724, USA
²UA Department of Surgery, Division of Surgical Oncology, 1501 N Campbell Avenue, Tucson, AZ 85724, USA
³NanoString Technologies, 530 Fairview Avenue North, Seattle, WA 98109, USA
*Corresponding author: Valentine Nfonsam, University of Arizona, Department of Surgery, Division of Surgical Oncology, 1501 N Campbell Avenue, AHSC 4334, Tucson, AZ 85724, USA
Received: June 24, 2016; Accepted: August 01, 2016; Published: August 03, 2016
Abstract
Background: Overall declines in incidence of rectal cancer (RC) in patients older than 50 years have been mostly attributed to improvement in treatment modalities and introduction of age-based screening. Recent studies, however, have shown a rise in the incidence of RC in patients younger than 50 years. The etiology of early-onset (EO) RC is not well understood. The aim of this study is to elucidate the molecular features of (EO) RC and show its uniqueness compared to late-onset (LO) disease.
Methods: Two cohorts of patients with sporadic RC were identified. Tumors and matching non-involved tissues from six (EO) RC patients (< 50 years) and six (LO) RC patients (>65 years) were obtained from Pathology archives. Deparaffinized tissues were macro-dissected from FFPE sections, RNA isolated and used for expression profiling of 770 cancer related genes representing 13 canonical pathways. Statistical analysis was performed using the Gene Expression R-script module within the nCounter software v2.6. A gene was considered to be above background if the average count for the target gene was greater than the average counts for the eight negative control genes and if the P value of the t-test was less than 0.05.
Results: When we compared rectal tumors to non-involved rectal tissues, changes in expression levels of 171 genes were statistically significant in earlyonset group and 151 genes in late-onset group. Further comparative gene expression analysis between early- and late-onset rectal tumors normalized to their matching non-involved tissues revealed that changes in expression of 65 genes were unique to early-onset rectal tumors with 16 genes being up- and 49 genes down-regulated using the cutoff criteria of expression levels difference >2 fold and p-value <0.01. At the pathway level, MAPK signaling was the most deregulated pathway in early-onset rectal tumors compared to PI3K-AKT signaling pathway being the most deregulated in late-onset rectal tumors.
Conclusions: Results of this study suggest that sporadic early-onset rectal cancer is characterized by distinct molecular events compared to late-onset disease.
Keywords: Sporadic early- and late-onset rectal cancer; Gene expression profiling; Pathway analysis
Introduction
Colorectal cancer (CRC) remains the third most commonly diagnosed cancer and the third most frequent cause of cancer related death in the United States with approximately one-third of patients with CRC having their cancer limited to the rectum [1-4]. Overall, CRC incidence and mortality rates are higher in men than in women [5] with higher percentage (31%) of rectal tumors being diagnosed in men compared to women (24%) [6]. In 2015, 23,200 new rectal cancer cases were predicted to be diagnosed in men and 16,410 in women in the United States [7]. There are also disparities in the age at which CRC is diagnosed with the median age at diagnosis being lower for rectal cancer (63 years in men and 65 years in women) and higher for colon cancer (69 years in men and 73 years in women) [8].
Even though there has been a significant progress in reducing the overall incidence of CRC over the past two decades, recent studies have reported an alarming increase in incidence within a population of CRC patients younger than 50 years [9-11]. Several studies have indicated a rise in colon cancer in younger patients; however, to the best of our knowledge, few have specifically addressed the incidence of cancer involving the rectum [10-13]. Meyer et al, in their 2010 study, analyzed the Surveillance Epidemiology and End Results (SEER) Program Cancer Registries data for incidence of rectal cancer between 1973 and 2005 and showed a significant increase in incidence of rectal and recto-sigmoid cancer in patients aged <40 years [13]. Our 2011 study demonstrated that the incidence of rectal cancer is continuously increasing in every age group (5-year intervals) from 20 years to 49 years, with the most impressive increase seen within the age group 40-44 years [11]. Our comprehensive 15-year analysis of the Arizona Cancer Registry revealed an alarming 225% increase in incidence of rectal cancer in the age group 30-34 years [14].
Overall declines in incidence of CRC in patients older than 50 years have been mostly attributed to improved treatment modalities and the introduction of age-based screening using fecal occult blood testing, sigmoidoscopy, or colonoscopy [15].
Biomarkers have been shown to be promising for cancer screening and diagnosis [16]. However, despite numerous significant technological and methodological advances, CRC research has not yielded a novel molecular biomarker or biomarker panel suitable for population-wide screening purposes [17]. Cytogenetic alterations such as microsatellite instability (MSI), chromosomal instability (CIN), and the CpG island methylator phenotype (CIMP) have been considered as potential CRC molecular markers as they can help with diagnostic, prognostic, and predictive treatment response information [18-20]. Molecular tests are expected to provide genetic information about the malignancy in progression. Intense research efforts aiming at identifying molecular markers (DNA, RNA, or protein) to develop novel, noninvasive biomarker detection methods for CRC in blood and stool are underway. Molecular markers can be used to assess the risk of malignancy, its aggressiveness over the time, and the probability that a patient will respond to a particular treatment what can result in helping physicians to make personalized treatment decisions. Even though there have been some promising advances in the use of biomarkers for detection of CRC, comprehensive molecular studies using larger cohorts of patients are necessary to determine if a single marker or a group of biomarkers can serve as first line test to diagnose CRC. The current conventional molecular tests used for evaluation of CRC patients include microsatellite instability (MSI) analysis and BRAF and KRAS mutation analysis. Innovative tumorbased molecular tests include CIMP, RNA expression, miRNAs and EGRF pathway biomarkers to predict response to anti-EGFR treatment in CRC patients with wild type KRAS [17].
The aim of this study was to perform a comprehensive genomic and pathway analysis of early- and late-onset rectal tumors in an attempt to answer questions about the etiology and uniqueness of early-onset rectal cancer and also look at possibilities for targeted and personalized treatments in this cohort of patients.
Materials and Methods
Patient samples
De-identified FFPE rectal cancer samples and matching noninvolved rectal tissues from a group of patients younger than 50 years (early-onset group) and a group of patients older than 65 years (lateonset group) were obtained from the University of Arizona Pathology archives. Patients with Lynch syndrome, familial adenomatous polyposis (FAP) and inflammatory bowel disease (IBD) were excluded. Patient samples were matched by stage of disease, gender and pathology. There was almost equal representation of males (n=2 for EO and n=3 for LO group) and females (n=4 for EO and n=3 for LO group), equal representation of stage II (n=2 for each age group) and stage III (n=4 for each age group) samples with the pathology of moderately differentiated adenocarcinoma for all samples (Table 1).
Samples
Age
TNM
Stage
Path
Gender
Race
neoadjuvant chemo
radiation therapy
RT before or after surgery
Surgical Procedure
EORC1
44
pT3N1cMx
III
Poorly differentiated to undifferentiated adenocarcinoma
F
White
yes
After
Laparoscopic total proctocolectomy
EORC2
41
pT3N1Mx
III
Moderately differentiated adenocarcinoma
M
Hispanic
After
Abdominoperineal resection
EORC3
47
pT3N0Mx
II
Moderately differentiated adenocarcinoma
F
Hispanic
Before
Low anterior resection
EORC4
48
pT3N1Mx
III
Moderately differentiated adenocarcinoma
M
White
After
Low anterior resection
EORC5
32
pT3N2aMx
III
Moderately differentiated adenocarcinoma
F
White
After
Low anterior resection
EORC6
50
pT3N0Mx
II
Moderately differentiated adenocarcinoma
F
White
After
Low anterior resection
LORC1
55
pT3N3Mx
III
Moderately differentiated adenocarcinoma
M
Native
yes
Before
Low anterior resection
LORC2
73
pT3N0Mx
II
Moderately differentiated adenocarcinoma
M
White
After
Abdominoperineal resection
LORC3
85
pT3N0Mx
II
Moderately differentiated adenocarcinoma
M
White
yes
Before
Low anterior resection
LORC4
63
pT2N1aMx
III
Well defferentiated to Moderately differentiated adenocarcinoma
F
Other
After
Low anterior resection
LORC5
80
pT2N1aMx
III
Moderately differentiated adenocarcinoma
F
Native
yes
Before
Low anterior resection
LORC6
83
pT3N2aMx
III
Moderately differentiated adenocarcinoma
F
White
yes
Before
Low anterior resection
Table 1: Clinical characteristics of rectal samples. Clinical data such as age at initial diagnosis, TNM, stage, pathology, gender, race; radiation treatment (RT), RT schedule (before or after surgery) and a type of surgical procedure are provided.
Tumor mismatch repair protein expression
One block of FFPE tumor tissue was selected per case and immunostaining was performed using standard protocols. Mouse anti- MLH1 (clone M1), anti-MSH2 (clone G219-1129), anti-MSH6 (clone 44) and rabbit anti-PMS2 (clone EPR3947) monoclonal antibodies from Cell Marque, Rocklin, CA were used for immunostaining. The intensity of immune-histochemical signal in the tissue sections was graded as (0), (1) weak, (2) moderate, or (3) strong. The proportion of positively stained cells was evaluated as a percentage. The score was calculated by multiplying the intensity and percentage of stained cells. Specimens were scored in a blinded fashion by a GI pathologist. A tumor was deemed negative for protein expression only if the neoplastic epithelium lacked nuclear staining, while non-neoplastic epithelial or stromal cells retain normal expression of that protein.
De-paraffinization, macro-dissection
Depending on tumor size, one to four continuous FFPE tissue sections (5 microns, mounted on positively charged slides) were required for nucleic acids isolation. The unstained tissue slides were incubated in series of three baths for 2 minutes each with gentle agitation for the first 15 seconds: d-limonene (histology grade), d-limonene, and 100% Ethanol. The slides were allowed to dry completely before re-hydrating in a 3% glycerol (MBG) solution. Using an Hematoxylin and Eosin (H&E) slide (taken continuously with the unstained sections) as a guide, a sterile razor blade was used to remove surrounding normal tissue from the unstained sections, and the remaining tumor tissue only was collected into a 1.7ml micro centrifuge tube.
Isolation of nucleic acids
Tumor tissue genomic DNA was extracted using the Maxwell® 16 FFPE Tissue LEV DNA Purification Kit (Promega, Madison, WI) according to manufacturer’s specifications. DNA yields varied from 2-50μg per case.
RNA isolation was performed using the Roche High Pure FFPET RNA Isolation spin-column kit according to manufacturer’s instructions. RNA was then quantified using the NanoDrop 1000 (Thermo Scientific), and its quality was assessed with the Bioanalyzer 2100 RNA Nano kit for Eukaryotic Total RNA (Agilent Technologies).
Tumor microsatellite instability (MSI) analysis
The panel of six mononucleotide markers (NR21, NR22, NR24, NR27, BAT25 and BAT26) [21] was used for multiplexed PCR amplification in tumor and matching non-involved normal tissue DNAs. PCR products were analyzed by capillary electrophoresis [22]. Tumors showing differences in marker-size between normal and tumor DNA at two or more loci out of six were classified as MSI, as described previously [22]. These cases were excluded from gene expression studies.
NanoString sample preparation and data analysis
100ng of the purified RNA was hybridized with the PanCancer Pathway Code Set (NanoString Technologies) at 65°C overnight. Further purification and binding of the hybridized probes to the optical cartridge was performed on the nCounter Prep Station, and finally the cartridge was scanned on the nCounter Digital Analyzer. RCC files from the NanoString Digital Analyzer were imported into nSolver2.6 software (NanoString Technologies) and were checked for data quality using default QC settings. All samples passed data quality QC. One out of six tumor samples from the early-onset group was excluded from further analysis due to observed discrepancies in expression changes when compared to other five samples within the same group. Background subtraction was carried out by subtracting the mean value of the eight NEG control ERCC sequences from the raw counts of all endogenous genes. All samples were normalized using the geometric mean of the housekeeper genes. Expression ratios were calculated by dividing the mean values of all samples in one experimental group, such as early-onset rectal tumor tissue, by the mean values of all samples in another experimental group, such as the matching non-involving rectal tissue. P-values were calculated by student’s t-test. The heat map in Figure 4 was generated using Multiexperimental Viewer 4.9.0. All other graphs in this manuscript were generated by nSolver2.6 and PanCancer Pathways Advanced Analysis module. Specifically, for the advanced analysis, we used the following arguments/parameters: “Type of data: raw; File type of plots: png; File type of plots: tiff; Low count threshold details: Remove Genes below Specified Threshold: TRUE; Threshold count value: 20; Remove genes below the threshold at frequency greater than: 0.5; Sample annotation details: Unique sample identifier: Sample. Name; Covariate1: Prognosis; Variable type: categorical; Reference level: normal; Normalization details: Perform normalization: TRUE; Auto-select number of housekeepers: TRUE; Pathway scoring details: Perform pathway scoring: TRUE; Pathway scoring method: PC1; Pathway scoring baseline variable: Progosis using normal; Plot pathway scores vs.: Progosis; Adjust pathway scores for: Differential expression analysis details: Perform differential expression testing: TRUE; Predictors: Progosis; Confounders: P-value adjustment: BY Run gene set analysis: TRUE; Path view details: Display results using Path view: TRUE; Color Path view path view plots by: Fold-change; P-value threshold: 0.05”.
Results
NanoString nCounter PanCancer Pathways Panel gene expression code set [23] was used to uncover differences in gene expression patterns between early-(patients younger than 50 years) and lateonset (patients older than 65 years) groups of patients with sporadic rectal cancer.
Gene expression profiling of sporadic rectal tumors
To access the deregulation of gene expression in both sporadic early-onset (age<50 years) and sporadic late-onset (age>65 years) rectal tumors, we employed NanoString PanCancer Pathways Panel gene expression analysis to quantify transcript levels of 770 genes representing 13 canonical cancer pathways [24-27]. Total RNA isolated from rectal tumors (N=6) and matching non-involved rectal tissues (N=3) was hybridized to the code set with approximately one million raw counts tallied for all genes in each sample. Raw data were processed and normalized using nSolver 2.6, following manufacturer’s guidelines (details in Materials and Methods). Five tumor samples and three control samples in early-onset age group and all tumor and control samples in late-onset age group were within technical ranges and used in further data analysis. Gene expression profiles were first analyzed for correlations among samples within as well as between experimental groups. As shown in Figure 1, there were positive linear correlations between tumor samples or between control samples, but not between tumor and control samples. Taken together, despite the limited patient sample sizes, the NanoString data sets were robust enough to reveal differences between tumor and matching noninvolved tissues at gene expression level.
Sporadic early-onset rectal tumors
PanCancer Pathways Advanced Analysis (PCPAA) was applied on early-onset rectal tumor data sets in terms to discover genes and pathways whose deregulation may underline changes in tumor physiology. The PCPAA module is an R-based statistical tool set that performs pathway-centered analysis of nCounter PanCancer Pathways Panel data. Among the 770 genes assayed, changes in expression levels of 171 genes were statistically significant between tumors and matching non-involved tissues (P<0.01, FDR<0.28). Among these, 61 genes had p-value less than 0.001 and FDR less than 0.08. The most statistically significant genes (p<0.0001) are listed in Table 2 (full list is provided in Supplemental Table 1), and also shown in a volcano plot (Figure 2A). Genes involved in a number of cancer pathways were enriched among the top differentially expressed genes. For example, genes in the MAPK signaling pathway appeared 7 times in the top 20 list (Table 2), followed by the RAS signaling pathway (6 times) and PI3K-AKT signaling pathway (4 times). Indeed, a pathway significance plot generated by the PCPAA module suggests that MAPK signaling pathway was the most deregulated in sporadic earlyonset rectal tumors (Figure 2B). Gene expression data were mapped onto KEGG pathway graphs by Path view function of the PCPAA, providing intuitive views of deregulation at the pathway level. A representative graph for the MAPK signaling pathway is shown in Supplemental Figure 1.
Log fold change
Lower confidence limit
Upper confidence limit
P-value
FDR
Pathways
HSPA2
-3.33
-3.68
-2.98
1.5E-06
0.01
MAPK
RASGRP2
-2.01
-2.32
-1.71
1.3E-05
0.03
MAPK, RAS
GNG7
-2.78
-3.25
-2.32
2.3E-05
0.03
PI3K, RAS
RXRG
-3.08
-3.62
-2.54
3.0E-05
0.03
TXmisReg
EFNA5
-2.4
-2.82
-1.98
3.0E-05
0.03
PI3K, RAS
CACNA2D2
-2.81
-3.31
-2.31
3.4E-05
0.03
MAPK
FLNC
-4.49
-5.41
-3.58
7.2E-05
0.03
MAPK
ITGA2
2.17
1.72
2.61
7.5E-05
0.03
PI3K
GRIA3
-2.22
-2.68
-1.77
7.6E-05
0.03
TXmisReg
MAP2K4
-0.899
-1.09
-0.713
7.9E-05
0.03
MAPK
HIST1H3H
1.96
1.55
2.37
8.3E-05
0.03
TXmisReg
PRKACA
-0.862
-1.04
-0.681
8.7E-05
0.03
Wnt, HH, MAPK, RAS, Apop
CCNB1
2.12
1.66
2.57
9.8E-05
0.04
CC
CACNA2D3
-2.68
-3.27
-2.09
1.1E-04
0.04
MAPK
SETBP1
-2.55
-3.11
-1.98
1.2E-04
0.04
RASA4
-1.41
-1.72
-1.09
1.2E-04
0.04
RAS
GNA11
-1.41
-1.73
-1.09
1.3E-04
0.04
CNTFR
-2.98
-3.69
-2.27
1.7E-04
0.04
STAT
BAD
-1.39
-1.72
-1.06
1.7E-04
0.04
PI3K, RAS, Apop
LIFR
-3.24
-4.05
-2.44
2.2E-04
0.05
STAT
Table 2: Most differentially expressed genes in early-onset rectal tumors. Top 20 most statistically significant differentially expressed genes between tumor and matching non-involved rectal tissues in patients 50 years old or younger. A gene is estimated to have 2^(log fold change) times its expression in baseline samples (non-involved rectal tissue). The 95% confidence interval for the log fold change is also presented, along with p-values and false discovery rates (FDR).
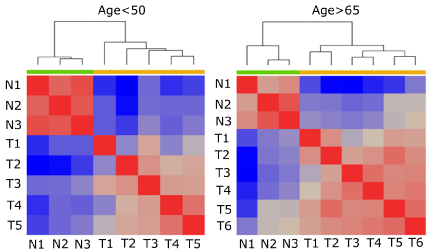
Figure 1: The Pearson correlation coefficient of gene expression between
rectal cancer and matching non-involved rectal tissue. Red indicates pairs
of samples with highly correlated gene expression profiles, grey indicates
correlations close to zero, and blue denotes highly negative correlations.
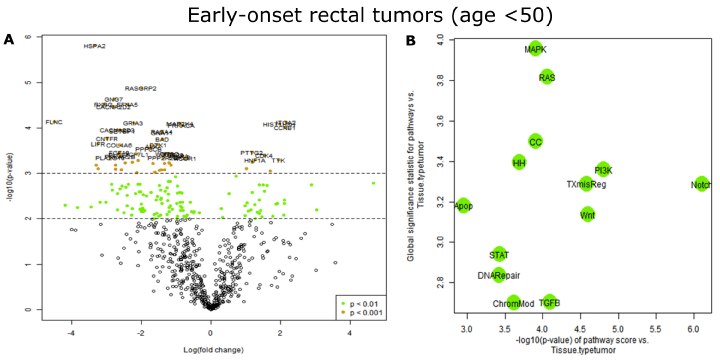
Figure 2: Differentially expressed genes in early onset rectal tumors. A). Volcano plot displaying each gene’s -log10(p-value) and log2 fold change with the
selected covariate. Highly statistically significant genes (P<0.001) fall at the top of the plot, and highly differentially expressed genes fall to either side. Horizontal
lines indicate thresholds for p=0.01 and p=0.001. The 40 most statistically significant genes (P<0.001) are named. B). Two measures of a pathway’s relationship
with a covariate are displayed. The first summarizes the pathways’ behavior in the differential expression analysis using their global significance statistics. A high
global significance statistic indicates a pathway’s genes are extensively differentially expressed in response to the covariate. The second measure summarizes
the behavior of the pathway scores with the covariate. For each pathway score, a linear regression has been fit predicting the pathway score from the selected
covariates. The horizontal axis plots each regression’s -log10(p-value) for the association between its pathway score and the covariate. A high -log10(p-value)
indicates a pathway score that is highly statistically significantly associated with the covariate.
Sporadic late-onset rectal tumors
PanCancer Pathways Advanced Analysis was also performed on late-onset rectal tumor data sets. Among the 770 genes assayed, changes in expression levels of 151 genes were statistically significant between tumors and matching non-involved tissues (P<0.01, FDR<0.35). Among these, 53 genes had p-value less than 0.001 and FDR less than 0.10. The most statistically significant genes are listed in Table 3 (P<0.0001) (full list is provided in Supplemental Table 2), and also shown in a volcano plot (Figure 3A). Genes known to be involved in the PI3K-AKT (7 genes), RAS (5 genes) and STAT (3 genes) signaling pathways were enriched among the top differentially expressed genes (Table 3). A pathway significance plot generated by the PCPAA module indicates that PI3K-AKT pathway was the most deregulated in sporadic late-onset rectal tumors (Figure 3B). Gene expression data were mapped onto KEGG pathway graphs to demonstrate deregulation at the pathway level. A representative graph of PI3K-AKT signaling pathway is shown in Supplemental Figure 2.
Log fold change
Lower confidence limit
Upper confidence limit
P-value
FDR
Pathways
COL11A1
4.93
4.32
5.53
9.3E-07
0.00
PI3K
GNG7
-2.44
-2.81
-2.08
3.6E-06
0.01
PI3K, RAS
RXRG
-3.22
-3.71
-2.73
4.1E-06
0.01
TXmisReg
FGF14
-1.36
-1.63
-1.09
2.4E-05
0.03
MAPK, PI3K, RAS
HPGD
-2.41
-2.89
-1.93
2.4E-05
0.03
TXmisReg
LIFR
-2.64
-3.18
-2.09
3.0E-05
0.03
STAT
SFRP4
4.36
3.43
5.28
3.6E-05
0.03
Wnt
GHR
-2.36
-2.89
-1.84
4.6E-05
0.03
STAT, PI3K
SOCS2
-1.49
-1.83
-1.16
5.3E-05
0.03
STAT
KIT
-1.97
-2.42
-1.52
6.2E-05
0.03
PI3K, RAS
INHBA
4.1
3.15
5.06
6.6E-05
0.03
TGFB
COMP
4.88
3.72
6.03
7.5E-05
0.03
PI3K
GRIN2A
-2.04
-2.53
-1.56
7.5E-05
0.03
RAS
GNAQ
-1.23
-1.52
-0.936
7.6E-05
0.03
RNF43
3
2.27
3.73
8.5E-05
0.03
VEGFA
1.77
1.34
2.2
8.5E-05
0.03
PI3K, RAS
MCM7
1.43
1.08
1.78
8.8E-05
0.03
CC
KLF4
-3.09
-3.87
-2.31
1.1E-04
0.03
APC
-0.937
-1.18
-0.696
1.2E-04
0.03
Wnt
RFC3
1.58
1.16
1.99
1.4E-04
0.04
DNA Repair
Table 3: Most differentially expressed genes in late-onset rectal tumors. Top 20 most statistically significant differentially expressed genes between tumor and matching non-involved rectal tissues in patients 65 years old or older. A gene is estimated to have 2^(log fold change) times its expression in baseline samples (noninvolved rectal tissue). The 95% confidence interval for the log fold change is also presented, along with p-values and false discovery rates (FDR).
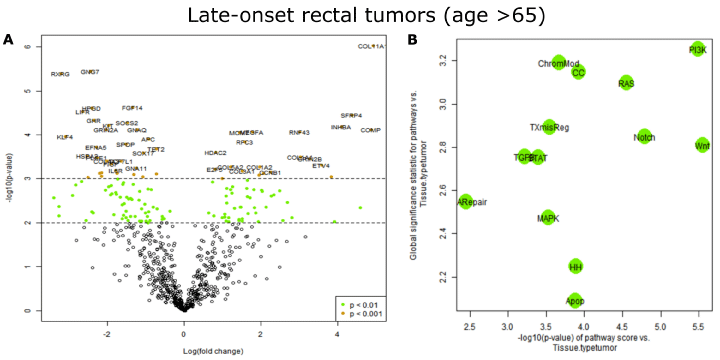
Figure 3: Differentially expressed genes in late onset rectal tumors. A). Volcano plot displaying each gene’s -log10(p-value) and log2 fold change with the selected
covariate. Highly statistically significant genes (P<0.001) fall at the top of the plot, and highly differentially expressed genes fall to either side. Horizontal lines
indicate thresholds for p=0.01 and p=0.001. The 40 most statistically significant genes (P<0.001) are named. B). Two measures of a pathway’s relationship with
a covariate are displayed. The first summarizes the pathways’ behavior in the differential expression analysis using their global significance statistics. A high
global significance statistic indicates a pathway’s genes are extensively differentially expressed in response to the covariate. The second measure summarizes
the behavior of the pathway scores with the covariate. For each pathway score, a linear regression has been fit predicting the pathway score from the selected
covariates. The horizontal axis plots each regression’s -log10(p-value) for the association between its pathway score and the covariate. A high -log10(p-value)
indicates a pathway score that is highly statistically significantly associated with the covariate.
Comparison of gene regulation in sporadic early- and late-onset rectal tumors
In order to address the critical question as to whether sporadic early-onset rectal cancer is a clinically and biologically distinct disease with characteristic molecular patterns and pathways driving its progression, we performed comparative gene expression analysis of sporadic early- and late-onset rectal tumors. The gene expression levels of the two age group rectal tumors were normalized to their matching non-involved tissues and clustered into a heat map to demonstrate an overview of similarities and differences in gene regulation (or rather deregulation) between the two age groups (Figure 4). Changes in expression of 21genes were statistically significant with a significance defined by difference of mean gene expression changes >2 fold and a p-value <0.05 (Supplemental Figure 3). Some subsets of genes were up- or down-regulated in both early- and late-onset rectal tumors, while other subsets were regulated uniquely in each of the two age groups. Using cutoff criteria of expression level difference >2 fold and p-value<0.01, 16 and 37 genes were only up-regulated in early-onset (blue box, Figure 5A) or late-onset (yellow box, Figure 5A) rectal tumors, respectively, while 21 genes were upregulated in both groups (green box, Figure 5A); 49 and 17 genes were significantly downregulated in early-onset (blue box, Figure 5B) or late-onset (yellow box, Figure 5B) rectal tumors, respectively, with 55 genes in common (green box, Figure 5B). At the pathway level, PI3K-AKT signaling was predominantly affected in late-onset tumors and MAPK pathway in early-onset tumors (Figure 2B and Figure 3B).
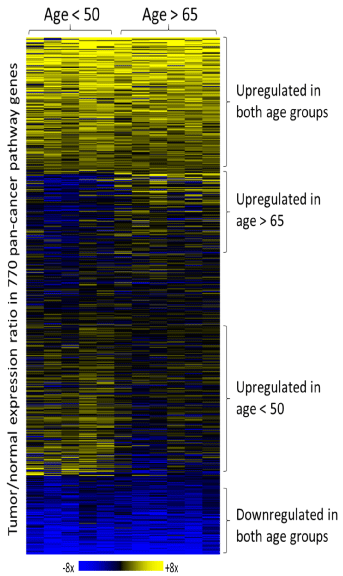
Figure 4: Overview of gene expression changes in early- and late-onset
rectal tumors. Changes in expression levels between tumor and normal
rectal tissues for 770 pan-cancer pathway genes are presented in a heat
map format. Color saturation represents the extent of the expression change,
with full saturation at 8-fold upregulation (yellow) or downregulation (blue).
Individual tumor samples are presented separately to illustrate reproducibility.
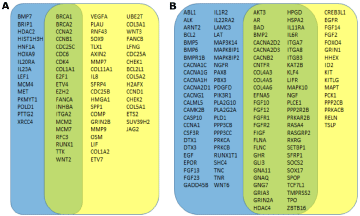
Figure 5: Differentially expressed genes unique to early- and late-onset rectal tumors. Mean gene expression levels in rectal tumor were compared to those in
matching non-involved rectal tissue. A). List of the most significantly up-regulated genes in early-onset tumors (blue box), late-onset tumors (yellow box) or in both
age groups (green box). B). List of the most significantly down-regulated genes in early-onset tumors (blue box), late-onset tumors (yellow box) or in both age
groups (green box). Significance is defined by difference of mean gene expression changes >2 fold and a p-value <0.01.
Discussion
A number of previous studies have suggested that the incidence of rectal cancer increased and continues to rise in patients whose age is below the recommended age-based screening [10-14]. These are typically patients younger than 50 years, not screened unless there is a family history of this type of cancer. Despite alarming increase in incidence of early-onset rectal cancer, the etiology is not well understood and there is still paucity in molecular data driving the early-onset rectal cancer initiation and progression. Our study was designed to uncover alterations in gene expression patterns and pathway mechanisms involved in sporadic early-onset rectal carcinogenesis and subsequent comparison with late-onset datasets.
The results from our study demonstrate that expression profiles differ significantly between sporadic early- and late-onset rectal tumors. When tumor tissues were compared to matching noninvolved tissues, unique sets of genes were found to be differentially expressed in each age group. Advanced pathway analysis revealed differences in pathways involved in early-onset and late-onset rectal carcinogenesis with PI3K-AKT signaling being the most affected in sporadic late-onset rectal tumors and MAPK and cell cycle signaling in sporadic early-onset rectal tumors.
Only a limited number of genomic studies that tried to uncover critical genes and pathways important in the initiation and progression of early-onset CRC are currently available. Our study adds to this growing body of literature and accentuates the fact that early- and late-onset rectal are two different entities. In our study expression analysis revealed that there are changes in expression of 65 genes that were unique to early-onset disease including genes such BMP7, LEF1, MET and RAD21.
Early-onset rectal cancers have been shown to be more aggressive [9,13,28]. In addition association between RAD21 expression and aggressiveness of CRC tumors and resistance to chemotherapy especially in KRAS mutant CRC tumors was recently reported [29] suggesting RAD21 as potentially important novel therapeutic target. Overexpression of RAD21 is seen in our study which might indicate why EO rectal cancer is more aggressive.
Previous studies have also suggested that expression of BMP7 in CRC and gastric tumors correlates with parameters of pathological aggressiveness such as liver metastasis, tumor recurrence and poor prognosis [30,31]. Our genomic analysis revealed BMF7 as one of the fifteen significantly up-regulated genes unique to early-onset rectal cancer. Thus, it is suggestive that BMP7 could serve as potentially useful clinical marker for young patients with rectal cancer.
Additionally, Lau et al. documented that the inappropriate activity of Wnt pathway in colorectal cancer can induce expression of LEF1 transcription factor which is normally not expressed in intestinal epithelium [32]. Another study by de la Roche et al. [33] documented that successful inhibition of oncogenic β-catenin in colorectal cancer requires the targeting of its interaction with LEF1. Moreover, overexpression of LEF-1 in colon cell lines was associated with increased proliferation in vitro and formation of neovasculature and size of tumor in vivo [34]. Our expression analysis revealed significantly up-regulated expression of LEF1 specifically in young patients with rectal tumors. Targeting of this gene can be potentially used for future development or applications of small-molecule inhibitors of oncogenic β-catenin in early-onset rectal cancers.
MET, gene found to be uniquely over-expressed in our early- onset rectal tumor samples, has already been demonstrated with its abilities to enhance colorectal tumor cell motility, facilitating invasion and metastasis [35]. Furthermore, it has previously been reported that MET is expressed in more than 50% of colorectal lesions from dysplastic adenoma to invasive carcinoma which is suggestive of its influence occurring from the early stages of malignant disease and its high association with advanced disease, worse prognosis and cancer-related mortality rates [36]. As MET was found to be uniquely expressed in early-onset rectal tumors, it too could be potentially considered as a valuable marker for poor prognosis and aggressiveness of early-onset rectal adenocarcinoma.
A few studies tried to uncover pathways playing significant roles in overall CRC carcinogenesis, including WNT, RAS-MAPK, PI3K, TGFβ, P53 and DNA mismatch-repair pathways [37]. However, studies looking at signaling pathways involved specifically in earlyonset rectal carcinogenesis are readily available. In our study, we demonstrate that MAPK and cell cycle signaling pathways are preferentially deregulated in sporadic early-onset rectal tumors while most of the genes that were differentially expressed in late-onset rectal tumors are involved in regulation of PI3K-AKT signaling.
This study strongly demonstrates that sporadic early-onset rectal cancer has unique molecular characteristics that are not seen in older patients. In addition, significant up-regulation of genes like BMF7, LEF1 and MET and RAD21 which seems to be uniquely involved in early-onset rectal carcinogenesis and survival, might explain why rectal cancers in young patients are more aggressive and confer poor survival. These findings of uniquely up-regulated genes specific to sporadic early-onset rectal cancer may indicate the promise of future studies using them as prognostic markers, small-molecule inhibitors of oncogenic pathways and development of potential less invasive diagnostic modalities. Furthermore, approaches targeting MAPK, WNT and cell cycle signaling could lead to the novel strategies and targets for treatment of sporadic early-onset rectal cancer, as well as new directions for the development of anticancer drugs in young patients with rectal adenocarcinoma.
Limitations of our study are the small sample size and that the samples are not equally matched for a treatment before and after surgery. Despite these limitations, our study strongly supports NanoString PanCancer panel as an effective platform for gene expression profiling of rectal cancer samples that are mostly available in FFPE format. Results of this study support our conclusions and corroborate with previous similar studies. We also know that more in depth studies of specific mechanisms of these findings are necessary to better understand the tumorigenesis, pathophysiology and also gain further insights into the molecular alterations of sporadic earlyonset rectal cancer.
References
- Myers EA, Feingold DL, Forde KA, Arnell T, Jang JH, Whelan RL. Colorectal cancer in patients under 50 years of age: A retrospective analysis of two institutions’ experience. World J Gastroentero. 2013; 19: 5651-5657.
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA: a cancer journal for clinicians. 2012; 62: 10-29.
- Siegel RL, Ward EM, Jemal A. Trends in colorectal cancer incidence rates in the United States by tumor location and stage, 1992-2008. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2012; 21: 411-416.
- Berry J, Bumpers K, Ogunlade V, Glover R, Davis S, Counts-Spriggs M, et al. Examining racial disparities in colorectal cancer care. Journal of psychosocial oncology. 2009; 27: 59-83.
- American Cancer Society. Colorectal Cancer Facts & Figures 2014-2016. Atlanta: American Cancer Society, 2014.
- Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA: a cancer journal for clinicians. 2014; 64: 104-117.
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015; 65: 5-29.
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, et al. Cancer statistics, 2008. CA: a cancer journal for clinicians. 2008; 58: 71-96.
- Kirzin S, Marisa L, Guimbaud R, De Reynies A, Legrain M, Laurent-Puig P, et al. Sporadic Early-Onset Colorectal Cancer Is a Specific Sub-Type of Cancer: A Morphological, Molecular and Genetics Study. Plos One. 2014; 9.
- O’Connell JB, Maggard MA, Liu JH, Etzioni DA, Livingston EH, Ko CY. Rates of colon and rectal cancers are increasing in young adults. Am Surgeon. 2003; 69: 866-872.
- Davis DM, Marcet JE, Frattini JC, Prather AD, Mateka JJL, Nfonsam VN. Is It Time to Lower the Recommended Screening Age for Colorectal Cancer? J Am Coll Surgeons. 2011; 213: 352-361.
- O’Connell JB, Maggard MA, Livingston EH, Yo CK. Colorectal cancer in the young. Am J Surg. 2004; 187: 343-348.
- Meyer JE, Narang T, Schnoll-Sussman FH, Pochapin MB, Christos PJ, Sherr DL. Increasing Incidence of Rectal Cancer in Patients Aged Younger Than 40 Years An Analysis of the Surveillance, Epidemiology, and End Results Database. Cancer. 2010; 116: 4354-4359.
- Nfonsam VN, Pandit V, DiGiovanni RM, Ohlson E, Aziz H, Jandova J, Gruessner AC. Increased Incidence of Early Onset Colorectal Cancer in Arizona: A Comprehensive 15-Year Analysis of the Arizona Cancer Registry. J Gastrointest Dig Syst. 2015; 5: 5.
- Edwards BK, Ward E, Kohler BA, Eheman C, Zauber AG, Anderson RN, et al. Annual report to the nation on the status of cancer, 1975-2006, featuring colorectal cancer trends and impact of interventions (risk factors, screening, and treatment) to reduce future rates. Cancer. 2010; 116: 544-573.
- Kumar S, Mohan A, Guleria R. Biomarkers in cancer screening, research and detection: present and future: a review. Biomarkers. 2006; 11: 385-405.
- Gonzalez-Pons M, Cruz-Correa M. Colorectal Cancer Biomarkers: Where Are We Now? Biomed Res Int. 2015.
- Lynch HT, de la Chapelle A. Genomic medicine - Hereditary colorectal cancer. New Engl J Med. 2003; 348: 919-932.
- Wheeler JMD, Loukola A, Aaltonen LA, Mortensen NJM, Bodmer WF. The role of hypermethylation of the hMLH1 promoter region in HNPCC versus MSI plus sporadic colorectal cancers. J Med Genet. 2000; 37: 588-592.
- Issa JP. Colon Cancer: It’s CIN or CIMP. Clinical Cancer Research. 2008; 14: 5939-5940.
- Buhard O, Cattaneo F, Wong YF, Yim SF, Friedman E, Flejou JF, et al. Multipopulation analysis of polymorphisms in five mononucleotide repeats used to determine the microsatellite instability status of human tumors. Journal of Clinical Oncology. 2006; 24: 241-251.
- Xicola RM, Llor X, Pons E, Castells A, Alenda C, Pinol V, et al. Performance of different microsatellite marker panels for detection of mismatch repairdeficient colorectal tumors. Jnci-J Natl Cancer I. 2007; 99: 244-252.
- Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008; 26: 317-325.
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000; 100: 57-70.
- Hanahan D, Weinberg RA. Hallmarks of Cancer: The Next Generation. Cell. 2011; 144: 646-674.
- Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004; 10: 789-799.
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou SB, Diaz LA, Kinzler KW. Cancer Genome Landscapes. Science. 2013; 339: 1546-1558.
- Chang DT, Pai RK, Rybicki LA, Dimaio MA, Limaye M, Jayachandran P, et al. Clinicopathologic and molecular features of sporadic early-onset colorectal adenocarcinoma: an adenocarcinoma with frequent signet ring cell differentiation, rectal and sigmoid involvement, and adverse morphologic features. Modern Pathol. 2012; 25: 1128-1139.
- Deb S, Xu H, Tuynman J, George J, Yan Y, Li J, et al. RAD21 cohesin overexpression is a prognostic and predictive marker exacerbating poor prognosis in KRAS mutant colorectal carcinomas. Brit J Cancer. 2014; 110: 1606-1613.
- Kazuo M, Fumiaki T, Yoshimasa K, Koshi M, Hiroyuki U, Hiroshi I, et al. Clinical significance of BMP7 in human colorectal cancer. Annals of Surgical Oncology. 2008; 15: 1530-1537.
- Aoki M, Ishigami S, Uenosono Y, Arigami T, Uchikado Y, Kita Y, et al. Expression of BMP-7 in human gastric cancer and its clinical significance. Brit J Cancer. 2011; 104: 714-718.
- de Lau W, Clevers H. LEF1 turns over a new leaf. Nat Genet. 2001; 28: 3-4.
- de la Roche M, Ibrahim AEK, Mieszczanek J, Bienz M. LEF1 and B9L Shield beta-Catenin from Inactivation by Axin, Desensitizing Colorectal Cancer Cells to Tankyrase Inhibitors. Cancer Res. 2014; 74: 1495-1505.
- Wang SH, Nan KJ, Wang YC, Wang WJ, Tian T. The balance between two isoforms of LEF-1 regulates colon carcinoma growth. Bmc Gastroenterol. 2012; 12: 53.
- De Oliveira ATT, Matos D, Logullo AF, Da Silva SRM, Neto RA, Longat A, et al. MET Is Highly Expressed in Advanced Stages of Colorectal Cancer and Indicates Worse Prognosis and Mortality. Anticancer Res. 2009; 29: 4807- 4811.
- Zeng ZS, Weiser MR, Kuntz E, Chen CT, Khan SA, Forslund A, et al. c-Met gene amplification is associated with advanced stage colorectal cancer and liver metastases. Cancer Lett. 2008; 265: 258-269.
- Fearon ER. Molecular Genetics of Colorectal Cancer. Annu Rev Pathol- Mech. 2011; 6: 479-507.