
Mini Review
Ann Hematol Oncol. 2019; 6(11): 1277.
MicroRNAs in Melanoma Resistance to Mitogen- Activated Protein Kinase Pathway Inhibitors
Letizia Motti M1,2*, Minopoli M2 and Carriero MV2
¹Department of Motor and Wellness Sciences, University “Partenope”, Italy
²Neoplastic Progression Unit, National Cancer Institute, Italy
*Corresponding author: Motti ML, Department of Motor and Wellness Sciences, University “Partenope”. Via Medina, 40. 80133, Naples, Italy
Received: October 31, 2019; Accepted: December 04, 2019; Published: December 11, 2019
Abstract
Background: The treatment of melanoma patients with MAPK pathway inhibitors is plagued by the development of drug resistance. Beside mutational events, recent studies highlight the notion that drug- resistance may be determined by altered microRNA (miRNA) expression in melanoma cells.
Objectives: The goal of this review is to provide recent updates on the mechanisms by which miRNAs regulate melanoma cell resistance to inhibitors of the MAPK pathway, including BRAF and MEK inhibitors
Methods: We conducted a literature review by keywords in the Pubmed and selected more recent articles
Results: miRNA deregulation appears to be one of the major responsible for the development of resistance to targeted therapies in melanoma. Emerging evidence shows that specific miRNAs are down- or up- regulated in drug resistant melanoma cells. In some cases, when expression of down-regulated miRNAs is restored, or alternatively, up-regulated miRNAs are silenced, a reversion of the resistant melanoma phenotype occurs both in in vitro and in vivo, confirming the central role of miRNAs in development of the drug resistance.
Conclusion: While studies in the miRNA field have grown exponentially in the last decade, the role of miRNA on the resistance to MAPK pathway inhibitors in melanomas is limited and much remains to be discovered. Understanding the mechanisms underlying miRNA-induced regulation of drug resistance in melanoma will represent in the future an important goal for the treatment of melanoma.
Keywords: Melanoma; Resistance to MAPK pathway inhibitors; miRNA; epigenetic modifications
Abbreviations
BRAFi: BRAF Inhibitor; MEKi: MEK Inhibitor; MAPKi: MAPK Pathway Inhibitor; miRNA: microRNA; CCL2: CC-Chemokine Ligand 2
Introduction
Melanoma is the most aggressive skin cancer, and its incidence has dramatically risen during the last fifty years [1]. Although combining targeted therapy and immune checkpoint inhibitors have improved significantly patient survival, effective treatments for metastatic melanoma are lacking to date, and the prognosis for these patients remains poor [2]. By next-generation sequencing, the Cancer Genome Atlas provided the analysis on the somatic aberrations underlying melanoma genesis, identifying BRAF, RAS, and NF1 mutant genetic subtypes of cutaneous melanoma all of them being able to deregulate MAPK/ERK pathway, leading to uncontrolled cell growth [3]. Over 50% of melanomas harbor activating V600E mutation in BRAF gene (BRAFV600E) which sustains proliferation and survival of melanoma cells by activating the Mitogen- Activated Protein Kinase (MAPK) pathway [4–8], whereas less common are substitutions of valine for lysine, arginine, leucine, or aspartic acid [9]. Inhibitors of BRAF-mutant specific kinase (BRAFi), such as vemurafenib and dabrafenib that inhibit the MAPK pathway, have been become worldwide standards of care for patients with BRAFmutant metastatic melanoma, improving their progression-free and overall survival [10,11]. However, their prolonged use is limited by early development of drug resistance and most of patients who initially respond to treatment with BRAFi, relapse within 6 to 8 months as a consequence of the activation of alternative proliferation-inducing pathways often associated to the reactivation of the MAPK pathway [12–16]. For these reasons the therapy for BRAF mutated melanoma has included the combination of different BRAFi with MEK inhibitors (MEKi) such as trametinib, cobimetinib or binimetinib) [17,18]. Although these combinations prolong overall and progression-free survival compared to single-agent therapies, resistance also occur in the majority of cases [18]. The scenario is complicated by the occurrence of PI3K/Akt upregulation, leading to BRAFi resistance in 22% of the melanoma patients [12]. Moreover, despite combination therapies targeting a variety of molecules, including Poli ADPribosio polimerasi inhibitors, have been employed to target different cellular pathways, most of them, do not escape development of drug resistance, due to the extraordinary plasticity of melanoma cells [19,20].
Non-durable therapeutic responses are mainly due to the high heterogeneity and plasticity of melanoma cells for the occurrence of genetic mutations as well as epigenetic modifications [21]. Indeed, drug resistance may be a consequence of transient adaptive resistance mechanisms. For instance, after exposure to MAPK pathway inhibitors, melanoma cells may undergo to different behaviors: a subset of cells undergoes apoptosis, a second subset remains arrested in the G0/G1 phase of the cell cycle (dormancy), and a third subset enter in a transient drug-resistant state by slowly cycling in an effort to minimize the effects of the drugs [22,23].
Emerging evidence assign to microRNAs (miRNA) an important role in regulating tumor pathogenesis, development and drug responsiveness [24–26] miRNAs are small non-coding RNAs of ~19–25 nucleotides that modulate gene expression by mRNA silencing or degradation, contributing to change cellular metabolism and genome stability. By targeting simultaneously multiple mRNAs, these epigenetic factors control a plethora of processes including cell proliferation and differentiation, cell senescence, survival, autophagy, migration and invasion [27]. Aberrant expression of miRNAs in melanoma cells compared to melanocytes is the result of chromosomal abnormalities, epigenetic regulation, and disorders in miRNA biogenesis [28–30] miRNA dysregulation has been observed during different stages of melanoma, and miRNAs are considered as biomarkers of melanoma progression with diagnostic and prognostic value [31–34]. It has been demonstrated that the MAPK signaling pathway, which is upregulated in melanoma, controls a network of 420 miRNAs [35] and recent studies highlight the notion that drugresistance may be determined by deregulation of a group of miRNAs. This review is focused to provide an updated overview of how some miRNAs influence melanoma cell resistance.
miRNAs as regulators of melanoma MAPKi-resistance
Several mechanisms involved in resistance to BRAF and MEK inhibitors have been identified to be modulated by miRNAs. Firstly, Liu et al, colleagues identified miR-200c as a pivotal signaling node in BRAFi- resistant melanoma cells for its ability to affect the MAPK and PI3K/AKT pathways, suggesting miR-200c as a potential therapeutic target for overcoming acquired BRAFi resistance. These Authors demonstrated that miR-200c inhibits drug resistance to PLX4720 BRAF and U0126 MEK inhibitors through downregulation of the p16 transcriptional repressor BMI-1, resulting in the inhibition of melanoma growth and metastasis formation in nude mouse xenografts. They also found that miR-200c acts on ABC transporters, a superfamily of transmembrane proteins that mediate drug resistance in melanoma cells [36]. The same authors confirmed the clinical significance of the miR-200c/Bmi1 axis in conferring acquired resistance to BRAFi therapy on human melanoma tissues. They showed that loss of miR-200c expression not only correlates with the development of resistance to BRAFi therapy in melanoma tissues, but also promotes development of a BRAFi-resistant phenotype in melanoma cells [37] (Figure 1). miR-514a-3p (miR- 514a), a member of a cluster of miRNAs on chrXq27.3, has been shown to have a role in the malignant transformation of melanocytes [38]. Stark and co-workers found that 69% of melanoma cell lines express a considerable amount of miR-514a that appear to be express in only 3% of other kind of solid tumors [39]. Using pull- down assay, the Authors showed that miR-514a binds to the NF1 transcription factor, inhibiting its expression. This results in the increased survival of PLX4032 (vemurafenib)-treated BRAFV600E melanoma cells. Moreover, the Authors demonstrated that a loss of NF1 correlates with a reduced BRAFi sensitivity of the melanoma cells [39] (Figure 1). The resistance to BRAFi may be partially reversed by the miR- 7. Using microarray profiling analysis of vemurafenib-resistant and parental A375 melanoma cells, Sun X. et al, colleagues found 17 dysregulated miRNAs in A375 cells resistant to BRAFi. Among these, miR-7 was identified as the most down-regulated miRNA in vemurafenib-resistant A375 melanoma cells. miR-7 inhibits the MAPK and PI3K/AKT signaling pathways and vemurafenib-resistant melanoma tumor growth in vivo by targeting EGFR, IGF-1R and CRAF [40] (Figure 1).
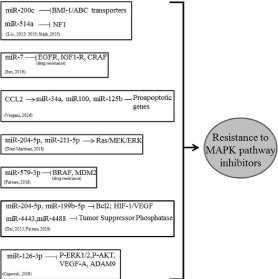
Figure 1: Schematic representation of most relevant miRNAs involved in
melanoma MAPK pathway inhibitors resistance.
By studying a panel of BRAFV600E melanoma cell lines with acquired resistance to BRAFi as well as plasma and tumor samples from vemurafenib-treated melanoma patients, Vergani and colleagues demonstrated that vemurafenib-resistant melanoma cells secrete higher levels of the CC-Chemokine Ligand 2 (CCL2) as compared to parental counterparts. CCL2 overproduction induces upregulation of miR-34a, miR- 100 and miR-125b which in turn negatively regulate apoptotic genes. Accordingly, inhibition of CCL2 and/or miRNAs silencing restores apoptosis, overcoming melanoma resistance to vemurafenib [41] (Figure 1).
More recently, Díaz-Martínez and co-workers, by the analysis of small RNAseq data and subsequent qPCR, documented increased miR-204-5p and miR-211-5p levels in vemurafenib-resistant A375 cells relative to parental counterparts. They found that cooverexpression of miR-204-5p and miR-211-5p stably induces Ras and MAPK upregulation after vemurafenib exposure. This effect has been detected not only in response to BRAFi but also in response to inhibitors of other elements of the MAPK pathway. In vivo, when transfected with both miR-204-5p and miR-211-5p, the parental A375 cells acquire vemurafenib resistance and high cell growth ability. Conversely, tumor growth was prevented by silencing resistant cells for miR-204-5p and miR-211-5p expression [42] (Figure 1).
In 2016 Fattore L. et al., colleagues given an important input to research studies on the relationships between miRNAs expression and drug resistance. They, demonstrated that miR-579-3p expression not only impairs the establishment but also reverts drug resistance to BRAF inhibitors. They found that miR-579-3p is down- regulated in vemurafenib-resistant melanoma cells and tissues from melanoma patients with acquired resistance to BRAFi. Consistently, low miR- 579-3p levels in tissues from melanoma patients correlates with a poor prognosis. The Authors found that miR-579-3p targets the 3’UTR of BRAF and MDM2, acting as oncosuppressor. Mechanistically, miR- 579-3p inhibits cell proliferation by targeting BRAF and increases apoptosis, by down-regulating MDM2 which results in p53 increase [43] (Figure 1). In 2019, the same group identified a large population of dysregulated miRNAs playing a role in development of drug resistance to BRAFi. They provided evidence that it is possible to block or revert development of drug resistance by regulating the expression of a subset of miRNAs. They presented evidence that transient overexpression of the two-downregulated miR-204-5p and miR-199b-5p in drug sensitive melanoma cells, causes inhibition of cell proliferation and induction of apoptosis. On the contrary, inhibition of the two upregulated miR-4443 and miR-4488 with specific antagomiRs, decreases the inhibitory effect of BRAFi on cell viability and induction of apoptosis. Interestingly, the Authors found that co-delivery of the down-regulated miR-204-5p, miR-199b-5p and miR-579-3p resulted in a moderate growth inhibition of A375 melanoma cells double resistant to BRAFi and MEKi, suggesting that the co-targeting simultaneously multiple microRNAs could be considered a valid approach to inhibit proliferation of double-drugresistant melanoma cells [44]. Finally, using matched tumor biopsies and serum samples from melanoma patients subjected to qRT-PCR to determine the expression levels of mir-4443, miR-4488, miR- 204b-5p and miR-199b-5p, the Authors identified specific miRNA signatures able to distinguish drug responding from non-responding patients [44] (Figure 1).
It has been reported that miR-199 impairs proliferative and proangiogenic HIF-1a/VEGF pathways [45]. Fattore L. et al., colleagues showed that down-regulation of miR-199 in BRAFi resistant melanoma cells, promotes VEGF release inducing angiogenesis, this phenotype being reverted by restoring miR-199 levels [44]. In line with these findings, Caporali and colleagues described the occurrence of a miRNA-dependent regulation of VEGF production in melanoma cells with acquired resistance to BRAF inhibitors. They found that miR-126-3p is down-regulated in dabrafenib-resistant melanoma cells as compared with their parental counterparts and that restoring miR-126-3p expression impaired proliferation and invasiveness of dabrafenib-resistant cells [46].
Conclusions
Malignant melanoma cells often develop resistance to most targeted therapies, including BRAFi and MEKi that inhibit the MAPK pathway. Mutations in the major driver genes, (BRAF, RAS, and NF1) are recognized to induce deregulation of the MAPK/ERK pathway, leading to uncontrolled cell growth. However, tumor resistance remains a therapeutic challenge since often, resistant tumors are lacking of genetic mutations. In recent years, melanoma cells have been shown to develop a transient drug adaptation by epigenetic control mechanisms. This is the case of miRNAs. As recapitulated in this review, miRNAs deregulation appears to be one of the the major causes for the development of resistance to targeted therapies in melanoma. When expression of specific miRNAs is restored in melanomas, reversion of the resistant phenotype is observed both in in vitro and in in vitro, confirming the central role of miRNAs in sustaining melanoma resistance. We believe that identification of pathways commonly deregulated by miRNAs in melanoma may lead to discover additional targets for therapeutic intervention.
Funding
MLM is funded by University of Naples “Parthenope” (DSMB 187, CUP I62I15000090005, 2017).
References
- Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics. CA Cancer J Clin. 2014; 64: 9–29.
- Sambi M, Bagheri L, Szewczuk MR. Current Challenges in Cancer Immunotherapy: Multimodal Approaches to Improve Efficacy and Patient Response Rates. J Oncol. 2019; 2019: 4508794.
- Zhang T, Dutton-Regester K, Brown KM, Hayward NK. The genomic landscape of cutaneous melanoma. Pigment Cell Melanoma Res. 2016; 29: 266–283.
- Akbani R, Akdemir KC, Aksoy BA, Albert M, Ally A, Amin SB, et al. Genomic Classification of Cutaneous Melanoma. Cell. 2015; 161: 1681–1696.
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002; 417: 949–954.
- Dhomen N, Marais R. BRAF signaling and targeted therapies in melanoma. Hematol Oncol Clin North Am. 2009; 23: 529–545.
- Fecher LA, Amaravadi RK, Flaherty KT. The MAPK pathway in melanoma. Curr Opin Oncol. 2008; 20: 183–189.
- Garnett MJ, Marais R. Guilty as charged: B-RAF is a human oncogene. Cancer Cell. 2004; 6: 313–319.
- Ascierto PA, Kirkwood JM, Grob J-J, Simeone E, Grimaldi AM, Maio M, et al. The role of BRAF V600 mutation in melanoma. J Transl Med. 2012; 10: 85.
- Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011; 364: 2507–2516.
- Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010; 363: 809–819.
- Shi H, Hugo W, Kong X, Hong A, Koya RC, Moriceau G, et al. Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer Discov. 2014; 4: 80–93.
- Van Allen EM, Wagle N, Sucker A, Treacy DJ, Johannessen CM, Goetz EM, et al. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov. 2014; 4: 94–109.
- Ascierto PA, Grimaldi AM, Anderson AC, Bifulco C, Cochran A, Garbe C, et al. Future perspectives in melanoma research: meeting report from the «Melanoma Bridge», Napoli, December 5th-8th 2013. J Transl Med. 2014; 12: 277.
- Palmieri G, Ombra M, Colombino M, Casula M, Sini M, Manca A, et al. Multiple Molecular Pathways in Melanomagenesis: Characterization of Therapeutic Targets. Front Oncol. 2015; 5: 183.
- Spagnolo F, Ghiorzo P, Queirolo P. Overcoming resistance to BRAF inhibition in BRAF-mutated metastatic melanoma. Oncotarget. 2014; 5: 10206–10221.
- Ascierto PA, Marincola FM, Atkins MB. What’s new in melanoma? Combination! J Transl Med. 2015; 13: 213.
- Moriceau G, Hugo W, Hong A, Shi H, Kong X, Yu CC, et al. Tunablecombinatorial mechanisms of acquired resistance limit the efficacy of BRAF/ MEK cotargeting but result in melanoma drug addiction. Cancer Cell. 2015; 27: 240–256.
- Fratangelo F, Camerlingo R, Carriero MV, Pirozzi G, Palmieri G, Gentilcore G, et al. Effect of ABT- 888 on the apoptosis, motility and invasiveness of BRAFi-resistant melanoma cells. Int J Oncol. 2018; 53: 1149–1159.
- Lord CJ, Ashworth A. PARP inhibitors: Synthetic lethality in the clinic. Science. 2017; 355: 1152–1158.
- Arozarena I, Wellbrock C. Phenotype plasticity as enabler of melanoma progression and therapy resistance. Nat Rev Cancer. 2019; 19: 377–391.
- Fallahi-Sichani M, Becker V, Izar B, Baker GJ, Lin J-R, Boswell SA, et al. Adaptive resistance of melanoma cells to RAF inhibition via reversible induction of a slowly dividing de-differentiated state. Mol Syst Biol. 2017; 13: 905.
- Ingangi V, Minopoli M, Ragone C, Motti ML, Carriero MV. Role of Microenvironment on the Fate of Disseminating Cancer Stem Cells. Front Oncol. 2019; 9: 82.
- Corrà F, Agnoletto C, Minotti L, Baldassari F, Volinia S. The Network of Noncoding RNAs in Cancer Drug Resistance. Front Oncol. 2018; 8: 327.
- Gelato KA, Shaikhibrahim Z, Ocker M, Haendler B. Targeting epigenetic regulators for cancer therapy: modulation of bromodomain proteins, methyltransferases, demethylases, and microRNAs. Expert Opin Ther Targets. 2016; 20: 783–799.
- Motti ML, D Angelo S, Meccariello R. MicroRNAs, Cancer and Diet: Facts and New Exciting Perspectives. Curr Mol Pharmacol. 2018; 11: 90–96.
- Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov. 2013; 12: 847–865.
- Bennett PE, Bemis L, Norris DA, Shellman YG. miR in melanoma development: miRNAs and acquired hallmarks of cancer in melanoma. Physiol Genomics. 2013; 45: 1049–1059.
- Mueller DW, Rehli M, Bosserhoff AK. miRNA expression profiling in melanocytes and melanoma cell lines reveals miRNAs associated with formation and progression of malignant melanoma. J Invest Dermatol. 2009; 129: 1740–1751.
- Wozniak M, Mielczarek A, Czyz M. miRNAs in Melanoma: Tumor Suppressors and Oncogenes with Prognostic Potential. Curr Med Chem. 2016; 23: 3136– 3153.
- Varamo C, Occelli M, Vivenza D, Merlano M, Lo Nigro C. MicroRNAs role as potential biomarkers and key regulators in melanoma. Genes Chromosomes Cancer. 2017; 56: 3–10.
- Mirzaei H, Gholamin S, Shahidsales S, Sahebkar A, Jaafari MR, Mirzaei HR, et al. MicroRNAs as potential diagnostic and prognostic biomarkers in melanoma. Eur J Cancer. 2016; 53: 25–32.
- Mannavola F, Tucci M, Felici C, Stucci S, Silvestris F. miRNAs in melanoma: a defined role in tumor progression and metastasis. Expert Rev Clin Immunol. 2016; 12: 79–89.
- Gajos-Michniewicz A, Czyz M. Role of miRNAs in Melanoma Metastasis. Cancers (Basel). 2019; 11.
- Couts KL, Anderson EM, Gross MM, Sullivan K, Ahn NG. Oncogenic B-Raf signaling in melanoma cells controls a network of microRNAs with combinatorial functions. Oncogene. 2013; 32: 1959–1970.
- Liu S, Tetzlaff MT, Cui R, Xu X. miR-200c inhibits melanoma progression and drug resistance through down-regulation of BMI-1. Am J Pathol. 2012; 181: 1823–1835.
- Liu S, Tetzlaff MT, Wang T, Yang R, Xie L, Zhang G, et al. miR-200c/Bmi1 axis and epithelial- mesenchymal transition contribute to acquired resistance to BRAF inhibitor treatment. Pigment Cell Melanoma Res. 2015; 28: 431–441.
- Streicher KL, Zhu W, Lehmann KP, Georgantas RW, Morehouse CA, Brohawn P, et al. A novel oncogenic role for the miRNA-506-514 cluster in initiating melanocyte transformation and promoting melanoma growth. Oncogene. 2012; 31: 1558–1570.
- Stark MS, Bonazzi VF, Boyle GM, Palmer JM, Symmons J, Lanagan CM, et al. miR-514a regulates the tumour suppressor NF1 and modulates BRAFi sensitivity in melanoma. Oncotarget. 2015; 6: 17753–17763.
- Sun X, Li J, Sun Y, Zhang Y, Dong L, Shen C, et al. miR-7 reverses the resistance to BRAFi in melanoma by targeting EGFR/IGF-1R/CRAF and inhibiting the MAPK and PI3K/AKT signaling pathways. Oncotarget. 2016; 7: 53558–53570.
- Vergani E, Di Guardo L, Dugo M, Rigoletto S, Tragni G, Ruggeri R, et al. Overcoming melanoma resistance to vemurafenib by targeting CCL2-induced miR-34a, miR-100 and miR-125b. Oncotarget. 2016; 7: 4428–4441.
- Díaz-Martínez M, Benito-Jardón L, Alonso L, Koetz-Ploch L, Hernando E, Teixidó J. miR-204-5p and miR-211-5p Contribute to BRAF Inhibitor Resistance in Melanoma. Cancer Res. 2018; 78: 1017–1030.
- Fattore L, Mancini R, Acunzo M, Romano G, Laganà A, Pisanu ME, et al. miR-579-3p controls melanoma progression and resistance to target therapy. Proc Natl Acad Sci USA. 2016; 113: E5005-5013.
- Fattore L, Ruggiero CF, Pisanu ME, Liguoro D, Cerri A, Costantini S, et al. Reprogramming miRNAs global expression orchestrates development of drug resistance in BRAF mutated melanoma. Cell Death & Differentiation. 2019; 26: 1267.
- Dai L, Lou W, Zhu J, Zhou X, Di W. MiR-199a inhibits the angiogenic potential of endometrial stromal cells under hypoxia by targeting HIF-1a/VEGF pathway. Int J Clin Exp Pathol. 2015; 8: 4735–4744.
- Caporali S, Amaro A, Levati L, Alvino E, Lacal PM, Mastroeni S, et al. miR- 126-3p down-regulation contributes to dabrafenib acquired resistance in melanoma by up-regulating ADAM9 and VEGF-A. J Exp Clin Cancer Res. 2019; 38: 272.