
Research Article
Austin J Med Oncol. 2025; 12(1): 1080.
On the Fidelity of Delta-Radiomic Models for Prediction of Pancreatic Tumor Response Following MRI-Guided SBRT
Hanson N, Dogan N, Simpson G, Spieler B, Jethanandani A, Mellon EA, Portelance L and Ford JC*
Department of Radiation Oncology, Sylvester Comprehensive Cancer Center and University of Miami Miller School of Medicine, Miami, FL, USA
*Corresponding author: John Chetley Ford, Ph.D., Sylvester Comprehensive Cancer Center, University of Miami Miller School of Medicine, Department of Radiation Oncology, 1475 NW 12th Avenue, Suite C123, Miami, FL 33136, USA Tel: 305-243-8895; Email: jcf137@miami.edu
Received: February 04, 2025; Accepted: February 21, 2025; Published: February 25, 2025
Abstract
Purpose: To ascertain the fidelity of predictive MRI-based delta-radiomics features in a moderately-sized cohort of pancreatic cancer patients.
Methods: MRI setup images from 37 patients treated to 50 Gy in 5 fractions on a 0.35T MR-Linac were subjected to radiomic analysis. Patients were classified as either responder (RS, n=17) or non-responder (NR, n=20) to treatment. The predictive power of radiomic and delta-radiomic features were examined using three feature selection algorithms, and logistic regression was used to build predictive models using the top 3, 2 and 1 feature for a total of 9 models. Patients were separated into a training set (n=25) and test set (n=12). The model building was repeated for expansions in the gross tumor volume (GTV) ranging from 0-10 pixels. Predictivity was measured via receiver operating characteristic Area Under Curve (AUC). The entire analysis was repeated, but replacing tumor response by a randomized outcome.
Results: Delta-radiomics was most predictive using relative change of ‘run-length nonuniformity’ feature at fraction 2. This was very consistent over the 9 models and most GTV expansions. A pronounced increase in predictivity using expansions of the GTV into the peritumoral region was noted. AUC in the training/test set was 0.85/0.75. Models built for the randomized outcome data appeared predictive for the training set but not in the test set (AUC = 0.69/0.50).
Conclusions: A multi-algorithm approach, along with multiple expansions of the GTV, and utilization of test set separate from the training set, is very useful in ascertaining the fidelity of radiomic predictive models.
Keywords: Radiomics; Delta-radiomics; Pancreatic cancer; MRI
Abbreviations
SBRT: Stereotactic Body Radiation Therapy; RS: Responder to chemoradiotherapy; NR: Non-Responder to Chemoradiotherapy; GTV: Gross Tumor Volume; ROC: Receiver Operating haracteristic; AUC: Area Under (the ROC) Curve; PDAC: Pancreatic Ductal Adeno Carcinoma; TRG-CAP: Tumor Response Grading with the College of American Pathologists; RF: Random Forest; LASSO: Least Absolute Shrinkage and Selection Operator; MRMR: inimum Redundancy Maximum Relevance; LOOCV: Leave-One-Out Cross Validation; AIC: Akaike Information Criterion; BED: Biological Equivalent Dose.
Introduction
Radiomics is the science of extracting quantitative features from medical images that may be subsequently exploited for prediction of patient outcome [1,2]. In the realm of oncology, with increasing attention toward personalized medicine [3], radiomics provides the potential to guide individualized medical management decisions. In the past decade, various researchers have utilized daily x-ray cone beam CT (CBCT) or magnetic resonance imaging (MRI) patient set-up images to examine changes in radiomic features during cancer treatment (delta-radiomics), resulting in promising models for predicting patient outcome [4-10]. However, a problem often encountered with radiomics analysis is overfitting; i.e., the high dimensionality of the potential radiomics feature space, which can number in the hundreds, is often large compared to the patient cohort number resulting in predictive features that are in fact only fitting to the statistical noise in the data rather than to any real underlying biological signal [11,12]. The aim of this paper is to ascertain the fidelity of predictive MRI-based delta-radiomics features in a moderately-sized cohort of pancreatic cancer patients.
Previous work by our group performed delta-radiomic analysis of the gross tumor volume (GTV) on 30 patients treated with low-field MRI-guided stereotactic body radiotherapy (SBRT) [10]. The analysis found two delta-radiomic features predictive of tumor response early during the treatment course with a receiver operating characteristic (ROC) area under curve (AUC) = 0.845, indicating a good predictor. However, due to the limited number of patients, only internal validation was feasible. We now have features extracted from 37 patients all treated with identical dose regimen and imaged identically and sought to repeat the delta-radiomic analysis with external validation, i.e., by splitting the cohort into training and test sets. We also sought to test the robustness of feature selection by utilizing multiple feature selection algorithms. Furthermore, we desired to understand the potential role of tumor contour variability, and whether expanding the volume of interest beyond the GTV would improve or affect the predictability. We also compared the tumor response prediction model to one where the outcome was a randomized binary outcome, to provide a baseline result in the absence of any real signal. Finally, we applied our model building tools to synthesized feature data, informed by our real data, to ascertain, in the presence of ground truth, how feature selection accuracy is affected by number of subjects.
Material and Methods
Patient Selection and Daily Setup MR Imaging
Patients in this study (N=37) had biopsy-confirmed pancreatic ductal adenocarcinoma (PDAC) and had completed chemotherapy prior to the MR-guided SBRT procedure on a 0.35T hybrid MRI/ radiotherapy unit (50 Gy in 5 fractions). A binary classification scheme identified patients as either responder (RS, n=17) or nonresponder (NR, n=20) to treatment. Treatment response for patients who had undergone curative-intent resection following SBRT utilized tumor response grading with the College of American Pathologists (TRG-CAP); TRG-CAP scores were considered responders and a score = 3 as NR. Response for remaining patients was determined with follow up dynamic CT, MRI and/or PET imaging studies acquired within 1-3 months after SBRT according to modified response evaluation criteria in solid tumors (mRECIST 1.1) [13,14]. Daily setup images were acquired using the clinical pulse sequence with 1.5x1.5x3mm voxels and nearly identical TR/TE (3ms/1ms) and bandwidth (540-600 Hz/pixel). All patients provided their written informed consent to participate in this study under an approved University of Miami Institutional Review Board protocol.
Radiomic Feature Extraction
GTVs on daily MRI setup images were contoured by a radiation oncologist with expertise in PDAC. Prior to feature extraction from the images, the intensity range of each GTV was normalized and quantized [10], and voxels resampled to 1.5mm isotropic. Radiomic features in the GTVs were calculated using the Texture Feature Toolbox in Matlab (Mathworks, Natick, MA). Features utilized in this work are listed in Table 1, along with shorthand codes for ease of reference. To account for contour uncertainty, and more importantly to explore whether important radiomic information exists outside the GTV, features were also extracted from eleven 1.5mm isotropic expansions of each GTV. Delta-radiomic features were calculated for fractions 2-5 according to (fxn - fx1)/abs(fx1), where fxn is the feature value for the nth fraction.
Encoding
MethodIBSI code/ aggregation code
Radiomic feature:
IBSI Name
Shorthand Code
GLCM
LFYI/IAZD
Energy
8ZQL
F1
Contrast
ACUI
F2
Entropy
TU9B
F3
Homogeneity
IB1Z
F4
Correlation
NI2N
F5
Sum Average
ZGXS
F6
Variance
UR99
F7
Dissimilarity
8S9J
F8
GLRLM
TPOI/IAZD
Short run emphasis
220V
F9
Long run emphasis
W4KF
F10
Gray-level nonuniformity
R5YN
F11
Run length nonuniformity
W92Y
F12
Run percentage
9ZK5
F13
Low gray-level run emphasis
V3SW
F14
High gray-level run emphasis
G3QZ
F15
Short run low gray-level emphasis
HTZT
F16
Short run high gray-level emphasis
GD3A
F17
Long run low gray-level emphasis
IVPO
F18
Long run high gray-level emphasis
3KUM
F19
Gray-level variance
8CE5
F20
Run length variance
SXLW
F21
GLSZM
9SAK/KOBO
Small zone emphasis
5QRC
F22
Large zone emphasis
48P8
F23
Gray-level nonuniformity
BYLV
F24
Zone-size nonuniformity
4JP3
F25
Zone percentage
P30P
F26
Low gray-level zone emphasis
XMSY
F27
High gray-level zone emphasis
5GN9
F28
Small zone low gray-level emphasis
5RAI
F29
Small zone high gray-level emphasis
HW1V
F30
Large zone low gray-level emphasis
YH51
F31
Large zone high gray-level emphasis
J17V
F32
Gray-level variance
BYLV
F33
Zone-size variance
3NSA
F34
NGTDM
IPET/KOBO
Coarseness*
-
F35
Contrast
65HE
F36
Busyness
NQ30
F37
Complexity
HDEZ
F38
Strength*
-
F39
*as defined by Amadasun and King [25].
Table 1: Radiomic features.
Feature Selection Algorithms and Predictive Model Building
The predictive power of radiomic features from fractions 1-5, the mean of features over fractions 1-5, as well as the delta-radiomic features, were examined using three feature selection algorithms: Random forest (RF) [15], Least absolute shrinkage and selection operator (LASSO) [16], and Minimum redundancy maximum relevance (MRMR) [17]. Patients were divided into a training set (11 RS/14 NR) and a test set (6 RS/6 NR). The three feature selection algorithms were applied to the training set to determine the top 3 radiomic and delta-radiomic features for each of the eleven GTV expansions. Logistic regression was then used to create predictive models of patient response using the top 3, top 2, and top 1 most predictive radiomic and delta-radiomic features from each algorithm for a total of 9 models for each GTV expansion, and AUC was calculated.
Model Comparison
Internal validation for each model was undertaken by performing leave-one-out cross validation (LOOCV). Another means of estimating the training set AUC and its uncertainty was undertaken by bootstrapping the data 1000 times performing logistic regression on 2/3 of the training set followed by application to the remaining 1/3, which afforded a mean AUC and 2.5 and 97.5 percentile confidence interval. The Akaike information criterion (AIC) [18], a widely used measure for predictive model comparison, was also calculated, as well as the accuracy for the training set. For external validation purposes, each logistic regression model was applied to the test set of patients, and AUC and accuracy of each model was calculated for the test set.
Further Analysis
The feature selection, model building, and model comparison described above was repeated but with patient outcome determined not by tumor response but classified randomly as positive or negative outcome. Care was taken to have balanced RS and NR in each class. In this way, we produce a data set with no possibility of a real predictive signal, only noise, for comparison to our tumor response data set that is hypothesized to contain a predictive signal (feature). Furthermore, for both the tumor response data set and randomized outcome data set, the training and test sets were resampled five times to investigate any dependence on training set/test set selection, taking care to maintain RS/NR balance in each. Finally, the delta-radiomic features from fraction 2 (and fraction 1) and GTV expansion of 4 pixels, were ranked by predictability based on Student t-test of difference in means between RS and NR. Synthetic data sets with varying numbers of subjects were created based on sampling of normal distributions using the mean and standard deviation, for RS and NR, of the top six features. These features were, along with p-value: F12, 0.001; F11, 0.002; F1, 0.069; F4, 0.098; F9, 0.113; F23, 0.118. Another 31 features were synthesized based on normal distributions with zero mean and standard deviation of 0.2, for both RS and NR. Synthesized feature data sets were created with patient number ranging from 10 to 300 and were subjected to feature selection by the three algorithms. This afforded the ability to test the accuracy of the feature selection algorithms in the presence of ground truth informed by our real patient data.
Results
We found overall the fraction 2 delta-radiomic features to be most predictive; therefore, all the results shown in this paper are for fraction 2 delta-radiomics. (For delta-radiomic results from all fractions, see the spreadsheet in the Supplemental Data) Shown in Table 2 are the results for Random Forest feature selection and tumor response prediction using the top 3, top 2, and top 1 features, for all eleven GTV expansions. F11 and F12 (Table 1 for shorthand codes) are consistently chosen for all models and expansions beyond two pixels. In the training set, bootstrapped estimation of AUC, LOOCV, conventional AUC, Accuracy, and AIC track with one another rather well and are consistent over the expansions beyond two pixels. (For AIC, lower value indicates more predictive, and differences of less than 2 are generally regarded as not significant.) AUC and accuracy in the test set show similar behavior, but with somewhat lower values compared to the training set.
Training Set
Test Set
Expansion (pixels)
Features chosen
AUC-BS (CI)
LOOCV
AIC
AUC
Accuracy
AUC
Accuracy
0
F22, F26, F11
0.55 (0.53 - 0.58)
0.52
36.49
0.53
0.56
0.58
0.58
1
F3, F12, F11
0.75 (0.72 - 0.78)
0.76
28.59
0.79
0.8
0.67
0.67
2
F11, F12, F14
0.80 (0.77 - 0.84)
0.84
27.99
0.91
0.92
0.58
0.58
3
F12, F11, F1
0.75 (0.72 - 0.78)
0.76
25.01
0.80
0.80
0.67
0.67
4
F11, F12, F16
0.75 (0.72 - 0.78)
0.72
30.21
0.84
0.84
0.42
0.42
5
F12, F11, F18
0.75 (0.72 - 0.78)
0.68
28.57
0.79
0.80
0.58
0.58
6
F12, F11, F23
0.71 (0.68 - 0.74)
0.72
27.52
0.84
0.84
0.75
0.75
7
F12, F11, F24
0.83 (0.80 - 0.87)
0.84
16.94
0.91
0.92
0.67
0.67
8
F12, F11, F34
0.75 (0.72 - 0.78)
0.72
28.63
0.79
0.80
0.42
0.42
9
F12, F34, F11
0.70 (0.67 - 0.73)
0.64
29.60
0.76
0.76
0.58
0.58
10
F11, F12, F21
0.80 (0.77 - 0.84)
0.76
29.87
0.95
0.96
0.58
0.58
0
F22, F24
0.58 (0.56 - 0.61)
0.56
34.53
0.53
0.56
0.58
0.58
1
F3, F12
0.75 (0.72 - 0.78)
0.76
26.79
0.76
0.76
0.58
0.58
2
F11, F12
0.75 (0.72 - 0.78)
0.64
28.69
0.79
0.8
0.75
0.75
3
F12, F11
0.75 (0.72 - 0.78)
0.72
25.74
0.76
0.76
0.58
0.58
4
F11, F12
0.73 (0.70 - 0.76)
0.72
29.49
0.83
0.84
0.67
0.67
5
F12, F11
0.75 (0.72 - 0.78)
0.76
30.19
0.79
0.8
0.83
0.83
6
F12, F11
0.75 (0.72 - 0.78)
0.76
27.22
0.79
0.8
0.67
0.67
7
F12, F11
0.71 (0.68 - 0.74)
0.72
31.95
0.7
0.72
0.75
0.75
8
F12, F11
0.75 (0.72 - 0.78)
0.76
28.39
0.79
0.8
0.67
0.67
9
F12, F34
0.71 (0.68 - 0.74)
0.72
28.22
0.72
0.72
0.67
0.67
10
F11, F12
0.70 (0.67 - 0.73)
0.64
32.89
0.74
0.76
0.83
0.83
0
F22
0.62 (0.60 - 0.65)
0.6
32.79
0.62
0.64
0.58
0.58
1
F3
0.75 (0.72 - 0.78)
0.72
32.4
0.75
0.76
0.5
0.5
2
F11
0.80 (0.77 - 0.84)
0.8
30.48
0.83
0.84
0.83
0.83
3
F12
0.75 (0.72 - 0.78)
0.72
27.57
0.78
0.8
0.75
0.75
4
F11
0.75 (0.72 - 0.78)
0.8
30
0.83
0.84
0.67
0.67
5
F12
0.75 (0.72 - 0.78)
0.76
28.62
0.78
0.8
0.75
0.75
6
F12
0.75 (0.72 - 0.78)
0.72
28.48
0.83
0.84
0.75
0.75
7
F12
0.75 (0.72 - 0.78)
0.72
29.97
0.74
0.76
0.75
0.75
8
F12
0.75 (0.72 - 0.78)
0.72
29.54
0.78
0.8
0.75
0.75
9
F11
0.71 (0.68 - 0.74)
0.68
32.67
0.75
0.76
0.83
0.83
10
F11
0.71 (0.68 - 0.74)
0.72
30.98
0.74
0.76
0.83
0.83
AUC-BS (CI) – bootstrapped AUC with confidence interval, LOOCV – leave-one-out cross validation,
AIC – Akaike information criterion, AUC – Area under curve.
Table 2: Random forest, fraction 2 delta-radiomics for predicting tumor response.
Shown in Table 3 are the results for Random Forest results, same as Table 2, but for randomized outcome prediction. Unlike the tumor response result, Table 3 shows no consistent feature selection over all models and GTV expansions. All else is similar to Table 2, but with generally lower predictability metrics especially in the test set.
Table 3: Random forest, fraction 2 delta-radiomics for predicting randomized outcome.
Table 4 summarizes the model results. It shows the top feature count (number of times a given feature is selected as the top predictor) and the mean AUC over all GTV expansions in the training set and test set for tumor response data as well as randomized outcome data, for all 9 models. For the tumor response data, it is seen that all nine models consistently choose F12 or F11 as the top feature. We also note that the best models according to the training set utilize 2 or 3 features, whereas according to the test set the three models using a single feature are more predictive. For the randomized outcome data, there is no consistency in top feature selection. AUCs in the training set are lower than for the tumor response data, but are still significantly positive (i.e., above 0.5), whereas the AUCs in the test set hover around 0.5 (not predictive).
Model
Tumor Response
Randomized Outcome
Top Feature Count
Training AUC
Test
AUCTop Feature
CountTraining AUC
Test
AUCF12
F11
Other
F12
F11
Other
RF 3
6
3
2
0.81 ± 0.11
0.59 ± 0.10
0
0
11
0.69 ± 0.05
0.61 ± 0.15
RF 2
6
3
2
0.75 ± 0.08
0.69 ± 0.09
0
0
11
0.66 ± 0.05
0.56 ± 0.13
RF 1
5
4
2
0.77 ± 0.06
0.73 ± 0.11
0
0
11
0.67 ± 0.07
0.61 ± 0.12
LASSO 3
9
1
1
0.85 ± 0.07
0.52 ± 0.08
0
0
11
0.69 ± 0.08
0.59 ± 0.17
LASSO 2
9
1
1
0.83 ± 0.07
0.59 ± 0.08
0
0
11
0.68 ± 0.05
0.58 ± 0.18
LASSO 1
9
1
1
0.74 ± 0.07
0.72 ± 0.08
0
0
11
0.66 ± 0.04
0.57 ± 0.13
MRMR 3
11
0
0
0.81 ± 0.07
0.57 ± 0.09
0
0
11
0.68 ± 0.06
0.48 ± 0.09
MRMR 2
11
0
0
0.82 ± 0.07
0.58 ± 0.08
0
0
11
0.67 ± 0.04
0.51 ± 0.13
MRMR 1
11
0
0
0.75 ± 0.06
0.73 ± 0.08
0
0
11
0.67 ± 0.04
0.52 ± 0.10
Highlights indicate best performing models.
Table 4: AUC (mean ± SD) over all GTV expansions for the 9 predictive models.
Figure 1 shows the behavior of the AUC averaged over all 9 models versus GTV expansion for the training set and test set for both the tumor response and randomized outcome data. AUC in the training set is substantially higher than for the test set, for both tumor response and randomized outcome data. Tumor response AUCs are significantly higher for expansions above 1 or 2 pixels as compared to no expansion, for both training and test sets. To test whether this behavior is a statistical coincidence, we re-plotted the AUC versus expansion for the tumor response data using 5 different assignments of patients to training and test sets. The behavior was consistent using all the patient reassignments. The randomized outcome data does not show this behavior. In Figure 1, test set AUCs appear to be predictive for the tumor response data, but not for the randomized outcome data. The test set AUCs for the randomized data appear to have a positive bias in Figure 1. To test this, we re-plotted the AUC versus expansion for the randomized data using 5 different assignments of patients to training and test sets. The mean test set AUC over all models and patient assignments was 0.50, demonstrating there is no AUC positive bias in the test set of the randomized outcome data.
Finally, Figure 2 shows the number of correctly selected predictive features for the three selection algorithms applied to the synthesized data. Random forest appears to be the best feature selector overall. It is seen that for > 100 subjects, all three algorithms choose at least 5 out of 6 predictive features. For 20 subjects, all three selection algorithms select at least the top 2 features.
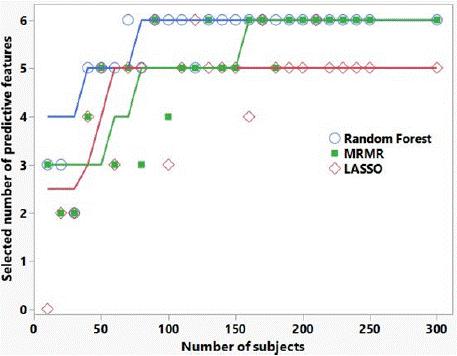
Figure 2: Number of correctly selected predictive features versus number
of subjects, for the three selection algorithms applied to synthesized data
informed by our patient feature data. There were 6 ‘predictive’ features, and
24 features with zero mean for both binary outcomes (i.e., not predictive of
outcome). The lines are Savitsky-Golay smoothing curves to guide the eye.
Discussion
Previous delta-radiomics work by our group [10] utilized lowfield MRI setup images in 30 PDAC patients treated to 30-60 Gy in 3-5 fractions. The non-uniformity of fractional dose led that study to perform delta-radiomics binned not by fraction, but by biological equivalent dose (BED). We found that the best predictor of tumor response was at 20 Gy BED, corresponding to either fraction 2 or 3 (normalized to fraction 1) delta-radiomics, with an AUC = 0.845. That previous work used internal validation only, and the predictive features were F6 and F37 (see shorthand codes in Table 1).
The current work differs from the previous study in that we utilized multiple feature selection algorithms, multiple regions of interest by expansions of the GTV, and external validation by separating the 37 patients into a training set and test set. In agreement with our previous work, the current study finds the best overall predictors of tumor response are observed early during treatment, using fraction 2 delta-radiomics. However, neither F6 nor F37, found predictive in the earlier work, are selected by any of the selection algorithms for any of the GTV expansions. This discrepancy highlights the uncertainty inherent in radiomic studies with limited patient cohort size and was the impetus for our desire to answer the question: Is there truly any information in the low-field MR setup images that may be predictive of pancreatic tumor response?
Part of the answer to this question may be addressed by noting the consistency of selected features over the 9 models and 11 GTV expansions (Table 2 and Table 4). Except for the 0-pixel and 1-pixel GTV expansions, all models/expansions chose F12 or F11 among the top predictive features. Other groups have explored the value of utilizing and comparing multiple radiomic models in predicting cerebral hemorrhage expansion [19] and for predicting tumor budding in rectal cancer [20]. Zhang, et al., demonstrated the utility of CT-based radiomics combined with multiple machine learning models to discriminate PDAC from pancreatic neuroendocrine tumor, and specifically recommended that future studies consider multi-algorithm modeling rather than a single algorithm [21]. The consistency of selected features from multiple algorithms in the current work lends credence to the multi-algorithm approach and lends some confidence that the features are predictive of tumor response.
Our initial motivation for performing delta-radiomics using expansions of the GTV was to account for contour uncertainty. Zhang, et al. [22], examined the effect of volume of interest (VOI) delineation on MRI-based radiomics to predict metastasis in nasopharyngeal carcinoma and sentinel lymph node metastasis in breast cancer, and found that smoothing the VOI or dilating by several pixels could improve radiomic analyses. Other researchers have reported on the importance of peritumoral radiomics since the region immediately surrounding the tumor parenchyma may be involved in immune infiltration, blood and lymphatic vasculature, and stromal inflammation ([23] and references therein). Takada, et al. [24], studied the ability of MRI-based radiomics to predict tumor control after radiotherapy in uterine cervical cancer, and found that an expansion of 4-8mm of the tumor VOI improved the AUC significantly. Hence, an additional motivation for performing deltaradiomics using expansions of the GTV in the current work was to determine whether there is radiomic information outside the GTV that would potentially improve prediction of tumor response.
Indeed, this study demonstrated (Figure 1) that tumor response predictivity improved by expanding the GTV by 1 or more pixels (>1.5mm), and this behavior was shown not to be a mere statistical coincidence of that particular sampling of patients into training and test set. This finding warrants further investigation, as discussed below. An additional benefit of performing radiomics in the GTV expansions is that it provides a degree of confidence in the robustness of the results. With limited patient cohort size, one is often uncertain whether an apparent positive radiomic result (i.e., a feature appears to be predictive of outcome) is merely overfitting to the noise in the patient feature data. GTV expansions provide multiple different noise sets. If a feature is found to be consistently predictive for the expansions, then this indicates there is real radiomic signal in the tumor and/or peritumoral region and is not due to overfitting to noise in a particular data set.
The AUC results shown in Table 4 and illustrated in Figure 1 highlight the importance of external validation. AUCs of the best performing models in the tumor response training set are approximately 0.85, consistent with our previous work. However, AUCs in the randomized outcome training set also appear to be predictive, when there is of course no real predictive radiomic signal in this case. Only by utilizing a separate test set do we see the AUC for the randomized outcome data go to approximately 0.5 (non-predictive) and see in the tumor response data the features are predictive (AUC = 0.73). Figure 1 also highlights the importance in this study of performing delta-radiomics in the GTV expansions. If we had only analyzed the GTV (expansion = 0) in our tumor response data, we would conclude that while the training set appeared to be predictive (AUC > 0.6), the test set indicated no predictivity of tumor response (AUC = 0.5); likewise, the randomly assigned data AUCs would indicate predictivity for both training and test sets (AUCs > 0.65 for GTV only). Finally, Figure 2 provides an indication of how patient cohort size affects the ability of the feature selection algorithms to find the top predictive delta-radiomic features, ranked by t-test discrimination of RS and NR, in our tumor response data. We cannot necessarily draw any strong conclusions about the relative utility of one selection algorithm versus another, as this is synthesized data informed by only one pancreatic tumor response data set; however, this sort of analysis may be beneficial in estimating numbers of subjects needed in future radiomic studies.
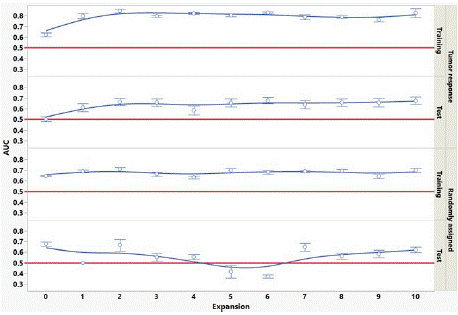
Figure 1: Mean AUC over all 9 models versus GTV expansion in training set
and test set, for tumor response and randomized outcome. Error bars are
standard error. Red lines at AUC=0.5 indicate no predictivity.
Conclusion
This study provides strong evidence of at least one MRI-based delta-radiomic feature able to predict tumor response in PDAC. The top predictive feature (in our shorthand denoted F12) is runlength nonuniformity from the gray-level run-length matrix. The relative change in F12 measured in low-field MRI setup images between fraction 2 (following only one SBRT fraction) and fraction 1 (a pre-radiotherapy scan) predicts tumor response in a test set of patients, independent of the training set, with an AUC = 0.75 using an expansion of at least 1.5mm of the GTV. This finding should motivate accrual of a separate patient data set for validation. The current work used only linear regression for model building but given confidence in the delta-radiomics results from this study and a future validation cohort, we may find that a different classification algorithm may improve the delta-radiomic feature’s predictive power. Additionally, this study’s finding that the radiomic signal appears to arise from the peritumoral area should motivate implementation of a technique to provide a map of predictive radiomic features throughout the GTV and surrounding tissue, to determine precisely from where the signal originates. This information could potentially allow not only MR imaging modifications to maximize the radiomic signal, but also, along with radiomics studies in pre-clinical models, give insight as to the pathophysiologic changes underpinning the delta-radiomics of pancreatic cancer.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
References
- Avanzo M, Stancanello J, El Naqa I. Beyond imaging: The promise of radiomics. Physica Medica. 2017; 38: 122-139.
- Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology. 2016; 278: 563-577.
- Hood L, Friend SH. Predictive, personalized, preventive, participatory (P4) cancer medicine. Nat Rev Clin Oncol. 2011; 8: 184-187.
- Fave X, Zhang LF, Yang JZ, Mackin D, Balter P, Gomez D, et al. Deltaradiomics features for the prediction of patient outcomes in non-small cell lung cancer. Sci Rep-Uk. 2017; 7: 588.
- Boldrini L, Chiloiro G, Casà C, Lenkowicz J, Carnero PR, Masciocchi C, et al. Predicting 2 years distant metastasis rate in rectal cancer: a MRI delta radiomics model. Radiother Oncol. 2019; 141: S36.
- Boldrini L, Cusumano D, Chiloiro G, Casà C, Masciocchi C, Lenkowicz J, et al. Delta radiomics for rectal cancer response prediction with hybrid 0.35T magnetic resonance-guided radiotherapy (MRgRT): a hypothesis-generating study for an innovative personalized medicine approach. Radiol Med. 2019; 124: 145-153.
- Cusumano D, Boldrini L, Yadav P, Casà C, Lee SL, Romano A, et al. Delta Radiomics Analysis for Local Control Prediction in Pancreatic Cancer Patients Treated Using Magnetic Resonance Guided Radiotherapy. Diagnostics. 2021; 11: 72.
- Delgadillo R, Spieler BO, Deana AM, Ford JC, Kwon D, Yang F, et al. Conebeam CT delta-radiomics to predict genitourinary toxicities and international prostate symptom of prostate cancer patients: a pilot study. Sci Rep-Uk. 2022; 12: 20136.
- Jin WH, Simpson GN, Dogan N, Spieler B, Portelance L, Yang F, et al. MRIbased delta-radiomic features for prediction of local control in liver lesions treated with stereotactic body radiation therapy. Sci Rep-Uk. 2022; 12: 18631.
- Simpson G, Jin W, Spieler B, Portelance L, Mellon E, Kwon D, et al. Predictive Value of Delta-Radiomics Texture Features in 0.35 Tesla Magnetic Resonance Setup Images Acquired During Stereotactic Ablative Radiotherapy of Pancreatic Cancer. Front Oncol. 2022; 12: 807725.
- Liu R, Gillies DF. Overfitting in linear feature extraction for classification of high-dimensional image data. Pattern Recogn. 2016; 53: 73-86.
- Gottardelli B, Gouthamchand V, Masciocchi C, Boldrini L, Martino A, Mazzarella C, et al. A distributed feature selection pipeline for survival analysis using radiomics in non-small cell lung cancer patients. Sci Rep-Uk. 2024; 14: 7814.
- Chatterjee D, Katz MH, Rashid A, Varadhachary GR, Wolff RA, Wang H, et al. Histologic grading of the extent of residual carcinoma following neoadjuvant chemoradiation in pancreatic ductal adenocarcinoma: a predictor for patient outcome. Cancer. 2012; 118: 3182-3190.
- Schwartz LH, Litière S, de Vries E, Ford R, Gwyther S, Mandrekar S, et al. RECIST 1.1-Update and clarification: From the RECIST committee. Eur J Cancer. 2016; 62: 132-137.
- Breiman L. Random forests. Mach Learn. 2001; 45: 5-32.
- Zou H. The adaptive lasso and its oracle properties. J Am Stat Assoc. 2006; 101: 1418-1429.
- Peng HC, Ding C, Long FH. Minimum redundancy - Maximum relevance feature selection. Ieee Intell Syst. 2005; 20: 70-71.
- Cavanaugh JE, Neath AA. The Akaike information criterion: Background, derivation, properties, application, interpretation, and refinements. Wires Comput Stat. 2019; 11.
- Duan CF, Liu F, Gao S, Zhao JP, Niu L, Li N, et al. Comparison of Radiomic Models Based on Different Machine Learning Methods for Predicting Intracerebral Hemorrhage Expansion. Clin Neuroradiol. 2022; 32: 215-223.
- Qu XT, Zhang L, Ji WN, Lin JZ, Wang GH. Preoperative prediction of tumor budding in rectal cancer using multiple machine learning algorithms based on MRI T2WI radiomics. Front Oncol. 2023; 13: 1267838.
- Zhang T, Xiang Y, Wang H, Yun H, Liu YC, Wang X, et al. Radiomics Combined with Multiple Machine Learning Algorithms in Differentiating Pancreatic Ductal Adenocarcinoma from Pancreatic Neuroendocrine Tumor: More Hands Produce a Stronger Flame. J Clin Med. 2022; 11: 6789.
- Zhang X, Zhong LM, Zhang B, Zhang L, Du HY, Lu LJ, et al. The effects of volume of interest delineation on MRI-based radiomics analysis: evaluation with two disease groups. Cancer Imaging. 2019; 19: 89.
- Tunali I, Hall LO, Napel S, Cherezov D, Guvenis A, Gillies RJ, et al. Stability and reproducibility of computed tomography radiomic features extracted from peritumoral regions of lung cancer lesions. Med Phys. 2019; 46: 5075-5085.
- Takada A, Yokota H, Nemoto MW, Horikoshi T, Matsushima J, Uno T. A multiscanner study of MRI radiomics in uterine cervical cancer: prediction of infield tumor control after definitive radiotherapy based on a machine learning method including peritumoral regions. Jpn J Radiol. 2020; 38: 265-273.
- Amadasun M, King R. Textural features corresponding to textural properties. IEEE Transactions on Systems, Man, and Cybernetics. 1989; 19: 1264-1274.