
Review Article
Austin J Nutr Metab. 2015; 2(4): 1026.
Genetic Variations in Vitamin D Metabolism Genes and the Microbiome, in the Presence of Adverse Environmental Changes, Increase Immune Dysregulation
Laing BB¹ and Ferguson LR1,2*
¹Discipline of Nutrition and Dietetics, FMHS, University of Auckland, New Zealand
²Auckland Cancer Society Research Centre, FMHS, University of Auckland, New Zealand
*Corresponding author: Lynnette R Ferguson, Auckland Cancer Society Research Centre, FMHS, University of Auckland, Auckland, New Zealand
Received: July 03, 2015; Accepted: August 06, 2015; Published: August 08, 2015
Abstract
Vitamin D metabolism in individuals is affected not only by their exposure to the sun or dietary intake of vitamin D, their liver and kidney function and other tissue production, but also by genetic variations in genes associated with vitamin D metabolism. These genes include not only the vitamin D receptor (VDR) genes, but also Group–Specific Component (GC), 7-dehydrocholesterol reductase (DHCR7), cytochrome P450 2R1(CYP2R1), cytochrome P450, family 24, subfamily A, polypeptide 1 (CYP24A1), and cytochrome p450 27B1 (CYP27B1). Particular single nucleotide polymorphisms (SNPs) from a number of these genes are being associated with lower or higher circulating vitamin D concentrations. Vitamin D metabolism is also affected by interactions with the individual’s microbiome. Other environmental changes in addition to vitamin D intake, particularly those associated with variations in nutrient intake and exposure to pathogens, can impact on these gene variations associated with vitamin D metabolism. Certain combinations of these, in addition to a lowering of circulating vitamin D, influence immune and metabolic pathways and contribute to immune dysregulation. The current enhancement and interplay of these permutations augurs the rise of chronic conditions in the Western world. To ameliorate this, vitamin D supplementation needs to be tailored to an individual’s genotype in the context of their environment.
Keywords: Genetic Variations; Vitamin D; Immune Dysregulation
Introduction
Vitamin D is a major regulator of gene expression and signaling in most tissues, and insufficiency affects many adults [1,2]. Genetic factors are thought to play a large part in this. Indeed, the genetic contribution to vitamin D status has been estimated to be from 28- 80% [3-5]. A number of particular single nucleotide polymorphisms (SNPs) for vitamin D associated genes or particular combinations of SNPs from genes relating to vitamin D metabolism can be involved in multiple signaling pathways. These pathways can be associated with lower or higher concentrations of circulating vitamin D. These particular SNPs from genes associated with vitamin D are also becoming increasingly relevant in their association with immune disorders.
Vitamin D deficiency is becoming well recognised as having a role in cell proliferation, differentiation, apoptosis and the immune response [6-13]. Vitamin D has been reported to be a significant factor in seventeen varieties of cancer [14-24], through its interactions with signaling pathways [8,21,25-31]. Vitamin D concentrations are also becoming particularly relevant in inflammatory disorders such as Crohn’s disease (CD), one of the common Inflammatory Bowel Diseases (IBD). Vitamin D plays a significant role in anomalous gene interactions, a critical factor in IBD [32-35]. Wang et al. showed that the active form, 1, 25 (OH) 2D3 affects the nucleotide-binding oligomerization domain containing 2 (NOD2) defense beta2 innate immune pathway, often found to be defective in people with CD [36]. Other immune disorders that have been associated with vitamin D are diabetes, coronary heart disease, blood pressure and outcomes of chronic kidney disease [37-49]. Particular variants of genes, the influence of microbiome, vitamin D metabolic pathways associated with them, and how our changing environment impinges on these and exacerbates immune dysfunction are the basis of the following discussion.
The Vitamin D Receptor Gene (VDR)
Most of the roles of the active form of vitamin D (1,25(OH)2D3) are facilitated by the gene VDR. VDR has a number of functions which include the following: it encodes the nuclear hormone receptor for vitamin D; it is a secondary bile acid lithocholic acid receptor which may act to protect the gut against lethal and carcinogenic endobiotics effects; it has a major role in calcium homeostasis; it affects the expression of 1a-hydroxylase (also known as cytochrome p450 27B1 (CYP27B1) which is associated with the conversion of the inactive form of vitamin D, 25(OH)D to the active form 1,25(OH)2D3; it increases the expression of 1,25(OH)2D3 which modulates cytochrome P450, family 24, subfamily A, polypeptide 1 (CYP24A1), a protein associated with the degradation of 1,25-dihydroxyvitamin. Mutations in this receptor can also lead to inherited rickets [50-55].
VDR is a major player among the genes involved with vitamin D metabolism because it influences so many other genes. As a member of the nuclear receptor family it directly modulates gene transcription, and is reported to influence the transcription of more than 913 genes [56]. Ramagopalan et al. found 2776 binding sites for VDR in their ChiP-seq defined genome wide map [57].
This influence of VDR on vitamin D concentrations in particular, is far reaching especially as it relates to immune regulation (Figure 1).
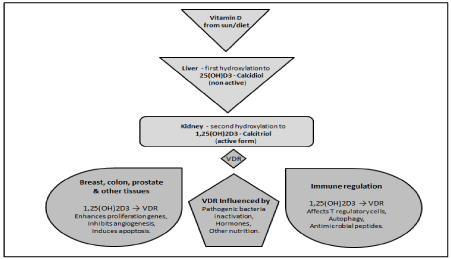
Figure 1: Vitamin D hydroxylation pathway to Calcitriol and the effect of VDR on tissues and Immune regulation.
Another example of VDR’s influences on immunity is its ability to bind to the fork head box P3 gene (FOXP3). This gene has a pivotal role in the maturity and performance of T regulatory cells [58]. Another role in which VDR is becoming increasingly significant is that of regulating autophagy. Autophagy is a process in animal cells by which malfunctioning cellular elements are transported to the lysosomes in the cell where they are deposed of or recycled. Wu et al. in their investigation on the effect of VDR deletion using cell culture models, VDR knockout mice, colitis models and human samples, found this led to defective autophagy in colitis [59].
The VDR gene, as part of the innate immune response, transcribes for antimicrobial peptides which are responsible for the deterrence and elimination of infection [60]. When the barrier function of the epithelium in the intestine is disturbed, this allows the microbes present to penetrate the intestinal epithelium, leading to inflammation. Metabolites of vitamin D act on anti-inflammatory pathways and are involved in maintaining the tight junctions between the epithelial cells of the intestine [61,62]. The gene protein tyrosine phosphatase, non-receptor type 2 (PTPN2) is also involved in the regulation of the epithelial barrier function, and is linked to CD [63]. If particular SNPs of the VDR gene impinge on this function, in combination with particular variants of the PTPN2 gene associated with CD, this would increase the risk of inflammation occurring.
Another factor linked to VDR and which may also contribute to the inflammatory process in conditions like CD, is the ability of some pathogenic bacteria to bind to and inactivate the VDR receptor. By doing this, they disarm the innate immune response and thus also increase the risk of inflammation. Gliding biofilm bacteria have been shown to produce a sulfonolipid capnine (2-amino-3-hydroxy- 15-methylhexadecane-1-sulfonic acid) which acts as a nanomolar kinetic inhibitor on the VDR ligand binding pocket [64,65]. Other examples of bacteria that are able to reduce the expression of VDR are Borrelia burgdorferi, Chlamydia trachomatis, Mycobacterium leprae and Mycobacterium tuberculosis [66-69]. Liu et al. showed in their work with Mycobacterium leprae that monocytes and macrophages can identify this bacterium, but the VDR response is impeded by Mycobacterium leprae upregulating the miRNA, hsa-mir-21 on the monocytes, and this mechanism allows them to avoid the vitamin D generated immune response [70]. The Epstein Barr virus has also been shown in vitro to down regulate VDR expression in B cells at mRNA and protein levels [71].
Studies on leprosy [68,70,72-74], considering how the disease interacts with the innate and adaptive immune systems, have also shown subtle variations in other key genes and enzymes associated with susceptibility or resistance to leprosy through VDR. These are NOD2, the mannose receptor, C type 1 (MRC1) and Leukotriene A4 hydrolase (LTA4H). When the host encounters Mycobacterium leprae, variations in the SNPs associated with these genes in combination can activate cytokine production through VDR and nuclear factor of kappa light polypeptide gene (NF-κB) receptor pathways.
Nutritional components other than vitamin D can also affect VDR and its activity. Curcumin, a polyphenol and component of the spice turmeric, is an example. Its’ active ingredient is diferuloylmethane, and it’s mechanism of action has been well studied [75-80]. It regulates many cell signaling molecules. Examples include: NF-κB, signal transducer and activator of transcription 3 (STAT3), hypoxia inducible factor 1, alpha subunit (HIF-1), peroxisome proliferatoractivated receptor gamma (PPAR?), arachidonate 5-lipoxygenase (5-LOX), tumour necrosis factor, (TNFa), interleukin 1, (IL-1) and interleukin 6receptor (IL-6) [76]. It can also attach to and activate VDR [77]. Curcumin therapy has been trialed in people with inflammatory disorders such as IBD, diabetes and cardiovascular diseases, where it has been shown to improve clinical symptoms [78- 80]. Curcumin is another factor involved in vitamin D metabolism, because by activating VDR it enables vitamin D concentrations to be enhanced.
VDR expression can also be moderated by hormones such as glucocorticoids, oestrogen, the parathyroid hormone and the retinoids. Glucocorticoids can affect the 25-hydroxyvitamin D3 24-hydroxylase (24(OH)ase), an enzyme involved in 1,25(OH)2D3 catabolism [81]. This means that when glucocorticoids are used as therapy, they can lead to excessive bone loss [82]. Oestrogen has been shown to up-regulate VDR via mRNA and VDR protein in tissues like the colonic mucosa, as illustrated by Smirnoff et al. [83]. Parathyroid hormone regulates VDR expression through VDR mRNA in the kidney and bone [17]. Retinoic acid increases the expression of VDR, especially in bone cells [84].
VDR effects on immune regulation are also influenced by its four commonly arising polymorphisms which have been variously linked with osteoporosis, bone mineral density, various cancers, heart disease, diabetes and CD. These polymorphisms are known as Fok1 (rs10735810- recently amalgamated with rs2228570 [85]) Bsm1 (rs1544410), Apa1 (rs7975232) and Taq1 (rs731236) [50,86].
The BsmI polymorphism has been suggested to have a significant association with bone mineral density in the lumbar spine [87] and with lower bone mass in patients with epilepsy [88]. However, other studies have not found these associations. In a recent systematic review on osteoporosis and VDR polymorphisms, 65.9% of studies reported a significant correlation between Bsm1 and osteoporosis risk, and 60.0% of studies reported a significant correlation between Fok1 and osteoporosis. Further research is needed to clarify the role of these polymorphisms, how these relate to participants’ vitamin D status and their relationship to other genetic and environmental factors, like ethnicity and gender, which can affect bone density [89- 91].
In a review of the literature with respect to cancer [86] Fok 1 has been most strongly associated with cancer risk for breast and prostate cancer and malignant melanoma. Bsm1 has been associated strongly with prognosis for breast cancer and malignant melanoma. Taq1 also has a strong association of prognosis for breast cancer and renal cell carcinoma [86].
Studies of VDR polymorphisms and coronary heart disease (CHD) show conflicting results particularly with respect to the VDR BsmI (B/b) polymorphism [92]. The B allele compared to the b allele has been associated with a predisposition to calcific aortic stenosis (n= 200), [93], and a higher intimal medial thickness on the common carotid artery (n=471), [94]. In another study bb was associated with severity of CHD (n=41), [95]. Contrary to this, in another investigation (n=3441), no associations were found with the prevalence and severity of CAD for either allele [96]. In a study on haemodialysis patients (n= 80) and the relationship of the BsmI polymorphism with left ventricular failure and atherosclerosis the BB genotype was associated with an increase in left ventricular mass and the authors suggested this could be used as a marker in these patients [44]. More recent work by Santoro et al. demonstrates the B allele maybe a genetic determinant for CHD and high blood pressure in chronic kidney disease (n=171) [46]. Studies on Fok1 and BsmI polymorphisms show mixed results with the risk of developing high blood pressure [40]. In the study by Hossein-Nezhad et al. on patients with coronary artery disease, their vitamin D concentrations and VDR polymorphisms were investigated. Those with at least one stenotic coronary artery had 25(OH)D serum concentrations < 10 ng/ mL and this was also associated with the FokI polymorphism [97].
Studies of Vitamin D polymorphisms associated with Diabetes (type 1 and 2) also show inconsistent results and a number of metaanalysis have been performed to find commonalities between studies [48,49,98-100]. In the meta-analysis by Wang et al. 79 studies were reviewed with respect to both types of Diabetes. From this analysis the Bsml polymorphism (B vs b) was associated with an increased risk of Type I Diabetes and the Folk 1 polymorphism (f vs F) with Diabetes Type 2 especially in those from East Asia. In the stratified meta-analysis, by Li et al. FokI polymorphism had a significant association with diabetic nephropathy (DN) in Caucasians [100]. In the study by Ferrareziet al. on Caucasian people an increased risk of CAD in people with type 2 diabetes was associated with a haplotype with the minor allele of Bsm1 and Taq1 and the major allele of Apa1 [101].
A meta-analysis performed in 2012 by Xue et al. [102] on nine studies on CD revealed a significant increase in risk for those carrying the Taq1 TT genotype (OR=1.23; 95% CI, 1.02-1.49). However, those carrying the Apa1 “a” genotype had a decrease in CD risk (OR =0.081; 95% CI, 0.67-0.97). A New Zealand (NZ) study based on a South Island cohort in Canterbury showed this to be true only for males [103]. Another more recent meta-analysis in 2014 by Wang et al. showed Taq1 was associated with an increased risk for IBD in an allelic comparison [104]. When a subtype disease, subgroup analysis was conducted this showed: a positive association for Apa1 with increased CD risk (AA versus aa); a significant association with BsmI in East Asian populations for IBD (BB plus Bb versus bb); for Taq1 in Caucasians (TT plus Tt versus tt) and a decreased risk with Folk 1 for IBD in Middle Easterners for all genetic types. The authors suggested that this may reflect that VDR polymorphisms have different roles in different ethnicities, a reflection that genetic susceptibilities for complex diseases are different for different indigenous groups. Vitamin D concentrations and their relationship to these polymorphisms may also differ with ethnic groups too.
In another NZ study, on CD and control subjects, based in Auckland, on a Caucasian population (n=608), Carvalhoet al. reported that the Taq1 genotype was significantly associated with CD and low circulating vitamin D concentrations (OR=0.893; 95% Cl, 0.825- 0.966). They also showed that low serum vitamin D concentrations had a significant association with the allele rs 73254 from the signal peptide, CUB domain, EGF-like 3 gene (SCUBE3) (OR= 0.917; 95% Cl, 0.847-0.992) in subjects with CD and in the control group [105]. Alleles associated with the SCUBE3 gene have been linked to renal cancer, hypermethylation and lung cancer [106-109].
A study by Levin et al. was conducted with white participants, who were followed up after a median 11 years on genetic variants and associations with 25(OH)D concentrations. They found with a lesser known SNP from the VDR gene (rs7968585), a low 25(OH) D concentration was associated with hazard ratios for the risk of the combined outcome of myocardial infarction, cancer, mortality and incident hip fracture for those who had 1 or 2 of the minor alleles [110].
VDR’s critical role has been further exemplified from studies with vitamin D receptor null mice. In the Bouillon et al. [111] review of research in this area, its’ significant role in calcium and bone modeling was very evident. In engineered VDR absent mice, a bone and growth plate phenotype emerged that was similar to humans with congenital rickets or serious vitamin D deficiency. Alopecia was another feature, as was the development of hypertension from high renin levels. Their immune system was also more sensitive. Autoimmune diseases like Diabetes type 1 and IBD were more likely to emerge if predisposing influences were in place. VDR null mice were also more likely to produce tumours when subjected to chemical carcinogens or oncogenes. Their cardiovascular system too was more vulnerable, with cardiac hypertrophy and thrombosis manifesting [111].
Other Gene Variations and their Links with Vitamin D Metabolism
SNPs associated with genes other than VDR also influence vitamin D levels, and a number of studies have shown their association with a higher risk of vitamin D insufficiency (Figure 2). Wang et al. in their GWAS study linked 15 cohorts (comprising about 30,000 people) from European descent to explore genes associated with vitamin D insufficiency. They showed in their study that a genotype score combining three variants from three different genes i.e. the SNP rs 2282679 from the gene Group –Specific Component (GC), the SNP rs12785878 from near the gene 7-dehydrocholesterol reductase (DHCR7), and the SNP rs10741657 near cytochrome P450 2R1(CYP2R1) increased the risk of vitamin D insufficiency [112].
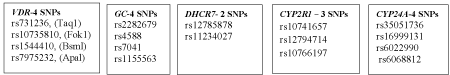
Figure 2: SNPs of genes associated with low circulating vitamin D concentrations.
Inflammatory conditions such as COPD, IBD and diabetes Type 2 have been associated with different variants in the GC gene [61,113,114]. The gene GC is the gene which encodes the vitamin D binding protein (VDBP) which is mostly formed by hepatocytes. VDBP is the main transport protein of vitamin D and its metabolites and is important for maintenance of the vitamin. VDBP has other significant functions such as fatty acid transport, the activation of macrophages and the actin scavenging system. Malnutrition may also lower its concentration [61,115-117]. Actin is released from cells upon injury, and GC binds actin to clear it from the circulation, thus preventing actin’s harmful effects on blood vessels [117]. Low plasma concentrations of VDBP may also be a result of particular polymorphisms in GC. Low levels of VDBP are an indicator of sepsis and organ failure following acute tissue injury [118]. Constans et al. noted in their study that the presence of GC la+ protein in patients with cirrhosis of the liver was associated with a poor outcome. They suggested that the presence of this protein in the sera may be a result of an aberration in the metabolism of VDBP. Moreover those who had low levels of 25-OH-D (2-7pg/l) did not show any change in VDBP with oral vitamin D3 supplementation [119].
The GC gene has over 120 variants but most of these are rare and relate to geographical location and skin colour. Three common alleles predominate (Gc1F, Gc1S and Gc2). These are defined by the genetic polymorphisms rs 4588 and rs 7041. People with white skins have a higher frequency of the Gc1S allele (50-60%) [120,121]. Higher levels of VDBP concentrations are also associated with Gc1S homozygote alleles as compared to Gc2 homozygote alleles [117,119,122,123]. According to the analysis by Wilson et al. there can be up to a 5 fold difference in the concentration of the vitamin D binding protein depending on the polymorphism. This, they suggest, may confound some studies. Perhaps like VDR polymorphisms, GC polymorphisms may also have different roles in different ethnicities. VDBP levels are not subject to seasonal variations but do have a diurnal rhythm [116,124]. An environmental factor like malnutrition may also affect VDBP concentrations.
The GC SNP identified in Wang’s et al. study has been identified in others [125,126]. Three other SNPs from the GC gene have also been associated with lowered vitamin D concentrations: rs4588, rs7041 and rs1155563 [126-129].
Vitamin D insufficiency also has links with genes associated with cholesterol. The DHCR7 gene encodes the enzyme involved with the last step of cholesterol synthesis where 7- dehydrocholesterol is converted to cholesterol. In humans, sunlight (ultra violet radiation 270-290 nm) converts 7- dehydrocholesterol to cholecalciferol - vitamin D3 [130]. People with low pigmented skin (i.e. with white skin) convert epidermal 7-dehydrocholesterol much more efficiently than those with a darker skin [131]. This has been linked to higher vitamin D status. This would confer a survival advantage to those living in Europe and North East Asia and is linked to the DHCR7 polymorphisms: rs12785878, rs7944926 and rs3794060 [132].
Cholesterol is also an important factor in the formation of bile acids which enable the absorption of dietary lipids [133]. It is possible that SNP rs12785878 may also be linked to the difficulty in digesting fats that people with CD report [134]. Another environmental influence on particular SNPs (rs1630498, and rs2002064,) for the DHCR7 gene (in the foetus) is the cholesterol level of their mother during pregnancy, which is linked to decreasing birth weight. Cholesterol serves as the main base for placental progesterone synthesis [135,136]. Over a hundred mutations in the DHCR7 gene are also associated with the Smith-Lemli-Opitz syndrome. This syndrome is associated with deformities in the palate, lungs, kidney heart, feet, and mental retardation [137,138].
To add to the complexity of this part of the vitamin D story, there is also a recent hypothesis that vitamin D concentrations may be influenced by genes involved with HDL cholesterol concentrations. Vimaleswaran et al. and Shirts et al. describe two SNPs (rs12272004 and rs3135506) from the Apolipoprotein A-V(APOA5) gene that interact with serum concentrations of 25(OH)D3 in the winter season, and this interaction affects HDL cholesterol [139,140]. The APOA5 gene has a significant role in the regulation of triglyceride levels in the plasma [115].
The third variant SNP which Wang et al. identified with vitamin D insufficiency is a member of the Cytochrome P450 super family rs10741657, associated with the CYP2R1 enzyme. This converts vitamin D into active ligands for VDR. This SNP of the CYP2R1 enzyme has been associated with lower 25(OH)D3 synthesis [115,141] and has also been detected in a growing number of other research studies on genetic influences on vitamin D concentrations involving mainly Caucasian groups [47,142,143]. Other rare mutants of this gene (L99P and K242N variants) have been associated with inherited rickets in non-Caucasian groups [144,145]. Teacher and colleagues in their study on this group with these variants noted that supplementation with vitamin D3 or D2 at 50,000IU/day led to low (in heterozygous subjects) to negligible rises (in homozygous subjects) in serum 25(OH)D, as compared to the rises in control subjects. The measurement of 25(OH)D2 and 25(OH)D3 used isotope-dilution liquid chromatography tandem mass spectrometry (LC-MS/MS) [144]. Mutair et al. found that 25(OH)D3 rose upon treatment with 5000 -10,000 IU/d of cholecalciferol(vitamin D3). The measurement taken was of total serum 25(OH)D, analysed using a competitive chemiluminescence immunoassay [145]. There are a variety of methods to measure serum vitamin D and debate about which is the best to use [146,147]. The difference in these results may reflect the different methodologies used to measure serum 25(OH)D.
In a recent study (n=92, Caucasians) by Nissen et al. conducted in winter time in Denmark, it was reported that, after the ingestion of vitamin D3-fortified bread and milk, carriers of the alleles rs10741657 (CYP2R1) and rs4588 (GC) as compared to non-carriers had the lowest concentrations and smallest increases in 25(OH)D concentrations. After 4 whole-body UVB treatments the carriers of the SNPs rs10741657 and rs4588 had the largest decreases in 25(OH) D concentrations after 6-months of consumption of vitamin D3- fortified bread and milk [148].
There is a hypothesis that vitamin D supplementation during the perinatal period maybe associated with the later development of allergy, as supplementation at this time suppresses dendritic cell development and later Th1 cell maturity [149-152]. As many western pregnant mothers and their infants are currently supplemented with vitamin D, an analysis on serum levels of 25(OH)D3 associated with gene variants was conducted on participants in the German Asthma Family Study (n=872, with white skin) to see if this was also a factor. This analysis showed at least one positive association with the transmission disequilibrium test of SNPs from 5 genes. (This test detects genetic linkage only when there is a genetic association presents [153].) This test included SNPs in the GC, CYP2R1 and CYP24A1 genes. With the SNP (rs10766197) from CYP2R1 there was an association with Asthma, IgE and 25(OH)D3 [154].
In a randomised control trial (n=1787, non-Hispanic whites) by Barry et al. across 11 centers in the USA with supplementation of vitamin D for a year (1000IU/day), it was found that the response to supplementation in 25(OH)D varied not only with respect to variants in CYP2R1, (e.g. rs10766197), but also to variants in Cytochrome P450, Family 24, Subfamily A, Polypeptide 1 (CYP24A1), (e.g.rs6013897) and VDR (e.g.rs7968585). The authors suggested that the results of their study possibly reflected that 25(OH)D is regulated by different mechanisms with respect to the source of vitamin D [155].
In a year-long study by Sollid and colleagues an association was found between the first measurement of serum 25(OH)D and BMI and carriers of SNPs associated with GC (rs2282679, rs7041) and CYP2R1 (rs10741657). Together these were a robust predictor of response to vitamin D supplementation of 20,000 IU of vitamin D3. Theauthors suggested that the implication of this is that some people would need higher doses of vitamin D supplementation to reach the recommended levels for serum 25(OH)D [156]. In a similar study by Waterhouse et al. in Australia, (n=350), 95% Caucasian, older adults (60-84 years) were administered 30,000 IU or 60,000 IU of vitamin D3 or a placebo over a year. From 84 SNPs tested from 41 candidate genes, they found the SNP (rs10766197) from the CYPR1 gene was associated with a significant change in circulating 25(OH)D level. They also reported that BMI, ambient UVR and self-reported health status had a small influence [157].
Another gene associated with the metabolism of vitamin D is the CYP24A1 gene which encodes the mitochondrial enzyme 1,25-dihydroxyvitamin D3 24-hydroxylase. This influences the levels of the active form of vitamin D (1,25(OH)2D3), especially its degradation. It also catalyses 25(OH)D and balances the action of the CYP27B1 gene which encodes the renal enzyme (P450c1a) involved in the second hydroxylation process to convert the inactive form of vitamin D (25(OH)D3) into the active form of vitamin D (1,25(OH)2D3). CYP24A1 is also influenced by the parathyroid hormone. This hormone, in the presence of 1,25(OH)D3, has been shown to up-regulate CYP24A1 in osteoblastic cells, suggesting it may have a role in preventing abnormal bone formation by modulating bone development [158]. Parathyroid hormone has also been shown in vitro studies to down regulate CYP24A1 in the kidney to avoid blood calcium levels fluctuating too widely [159]. Fibroblast growth factor 23 (FGF23), a protein encoded by the FGF23 gene (which is responsible for phosphate metabolism), also regulates the expression of CYP24A1 in the kidney and thus lowers serum 1,25(OH)2D3 levels [160].
Currently the CYP24A1 gene has around 50 polymorphisms ascribed to it. Four of these have been associated with lower concentrations of vitamin D, but little is known about many of the others [159]. Some of them have been identified with specific population groups e.g. African Americans. In a study by Roff and Wilson on this population, eight SNPs were identified as unique to African Americans [161]. One of these SNPs associated with the promoter CYP24A1 region was shown to reduce the expression of CYP24A1 in cultured human lymphocytes in vivo [161]. Another SNP has been recently detected as being associated with idiopathic infantile hypercalcemia [162]. In the study by Jacobset al. on vitamin D metabolism and cancer cells, four SNPs in this gene lowered CYP24A1 enzyme activity [163]. Future research will identify whether the other polymorphisms of CYP24A1 can be linked to immune dysregulation with other inflammatory disorders such as cancer, CD and heart disease. In the German Asthma Family Study, the authors noted a 5-point frequent haplotype with CYP24A1 (rs2296241, rs17219315, rs2762942, rs2248137, rs2248359), which was associated with a diagnosis of asthma, total IgE, 25OH2D3 and 1,25-OH2D3 serum concentrations [154].
The CYP27B11 a-hydroxylase, encoded by the CYP27B1 gene, is associated with the conversion of the inactive form of vitamin D, 25(OH)D, to the active form 1,25(OH)2D3. Mutations in this gene are mainly linked with vitamin D-dependent rickets [164]. Up to 38 mutations relating to this disorder have been identified [165]. A number of other variants in this gene are beginning to be associated with lower (25(OH)D levels and particular disorders e.g. multiple sclerosis (rs12368653, rs10876994, rs118204009, rs703842,andrs4646536) [166-170], and gestational and type 1 diabetes (rs10877012) [171,172] in Caucasian populations. Some of these SNPs are also found in other ethnic groups such as African Americans and Han Chinese [125,173]. In the study by Jacobs et al. on vitamin D metabolism in cancer cells, four SNPs (rs28934604, rs58915677, rs2229103 and rs13377933) were associated with reduced enzyme activity and one (rs8176344) increased enzyme activity compared to a wild type control [163]. The authors comment that these changes would result in varying exposure to 1,25(OH)2D3 in colonic cells, which could make people more susceptible to colon cancer developing. This suggested to them that vitamin D intake would need to vary according to genotype to maintain optimal vitamin D metabolite concentrations in the circulation [163].
The Influence of the Microbiome
The microbiome is the genes and genomes of the microbiota, a large collection of diverse bacterial, archaeal, and eukaryotic microbes which form an intricate ecosystem within different habitats of the human body [174-176]. Each body part has its own microbial composition irrespective of age, gender, although metagenomic data indicate that the metabolic pathways are essentially the same across all body sites [177]. The microbiome in the human gut, for example, contains up to (1014) microbes with a density about 1011 -1012 cells/ ml which encompasses over 3 million non-redundant genes. This implies that compared to the human genome, the microbiota have 150 times more genes they can access [178,179]. The microbiome is also genetically alterable which is dissimilar to the human genome. The microbiome alters as microbiota change with fluctuations in the environment and to variable food intakes [180]. The microbiota also influences the immune system e.g. their metabolites limit histone deacetylases to stimulate regulatory T cells, and they have a pivotal role in maintaining the integrity of the gut epithelium and its barrier function [181-184]. Microbiota dysbiosis has been linked to IBD, obesity, type 2 diabetes, and cancer [185-190].
Studies are revealing that vitamin D concentrations and VDR have an effect on the microbiome, and hence the genes and genomes there. This needs to be considered when discussing immune dysregulation and vitamin D.
A study by Jin and colleagues compared caecum contents with faecal samples from whole-body VDR knockout and wild-type mice. They found that there were differences in how bacterial contents changed in the transition from the caecum to the faeces between the two types of mice. This, they suggested, could lead in VDR knockout mice to dysbiosis and altered functions of the gut microbiome [191].
Chen et al. studied VDR knockout mice and their colonisation resistance to Citrobacter rodentium [192]. This is a gram-negative enteric bacterium found in mice, which causes colonic hyperplasia. This bacterium shares a number of pathogenic mechanisms in common with two significant human gut pathogens: Escherichia coli (EPEC) and Enterohaemorrhagic coli (EHEC). The data suggested that VDR not only influences the colonisation and vulnerability of the host to these bacteria, but has an effect on gut microbiota through the regulation of Innate Lymphoid Cell (ILC3) numbers and function. Chen et al. suggested that interventions with vitamin D could help control dysbiosis by normalizing ILC function [192].
There are also other pattern recognition receptors which are regulated by vitamin D and have an influence on the microbiota. For example, NOD2, in recognising bacteria peptidoglycans can cause the destruction of the bacteria through autophagy, stimulating antimicrobial production [36,193-196]. Vitamin D is also used by macrophages and dendritic cells to stimulate the production of antimicrobial peptides like B-defensin and cathelcidin. Where there is a wound, 1,25(OH)3 increases expression of the genes coding for the microbial pattern recognition receptors CD14 and TLR2 [36,197]. Thus, these genetic pathways associated with vitamin D and the microbiome may contribute to immune function or dysfunction depending on the genetic variation present.
The type of first foods also contributes to the composition of the microbiota. Breast feeding promotes immune development and protects against many diseases. In a small study on 3 month old babies 1,214 probe sets for epithelial cells were significantly, differently expressed in exclusively breast fed (n= 12) and formula fed babies (n=10) [198]. This may explain some of the differences that have been noted in clinical and epidemiological observations between these two groups. The two groups have different gut development with the breast-fed babies being described as being ‘less leaky’ [198]. In work done by Donavan et al. on piglets, the analysis of the stools of the microbiome gene expression at Phyla level showed those that were all formula fed shared the same distinct signature, but those that were suckled showed highly variable signatures. Bacteroides species, which are important players in the commensal bacteria, were not found in the stools of the formula fed piglets [198]. The microbiome of those that were suckled also had a higher number of virulence genes, and this was thought to contribute to the host immune defense genes. These observations could help explain why babies who are breast fed have a gut that is less leaky. Breast milk also contains human milk oligosaccharides (HMOs), which stimulate the growth of the intestinal flora, acting as prebiotics for beneficial bacteria [199]. This also would contribute to the protection of the gut barrier function.
Another element which can intrude on the microbiota working together to protect the intestinal barrier function is exposure to antibiotics. Although the microbiota exhibit resilience to antibiotics, repeated exposure can induce an importunate regime shift which can lower resistance. In the study by Dethelsen and Relman healthy subjects who had not had antibiotics in the last year were exposed to two standard five day courses of the antibiotic Ciprofloxacin, at six month intervals. Their analysis of the microbiota community in the faecal output, after the completion of these courses, showed a large loss of diversity and change in composition of the subjects’ microbiota community. Recovery of the microbiota community began seven days after the last dose of antibiotics. However, it was not always to the previous level before treatment began. The authors suggest that each course of antibiotics may shift the microbiota community from mutualistic patterns and make them more vulnerable to pathogenic strains of microbiota and enable the intestinal barrier to be penetrated [176,200]. How these shifts were also affected by variants in VDR and the other genes involved in vitamin D metabolism could also be a contributing factor in recovery.
Conclusions
Vitamin D metabolism is influenced not only by lack of exposure to sunlight or food sources of vitamin D, the liver and kidney function, and other tissue production, but also an individual’s genetic identity, their microbiome and environment. An individual’s genes (including microbial genes) can have a strong influence on metabolic pathways associated with vitamin D, and this can impact on immune regulation. Dysfunctional polymorphisms of genes, acting autonomously or together, environmental influences such as disabling of genes by pathogens, changes in nutritional status, and increasing antibiotic use added to insufficient exposure to vitamin D, all fuel aberrant vitamin D metabolism. These collectively contribute to immune dysregulation and to the rise in chronic conditions in the Western world. To impede this trend, vitamin D supplementation needs to be tailored to an individual’s genetic variants in the context of their environment.
References
- Holick MF. Diabetes and the vitamin d connection. Curr Diab Rep. 2008; 8: 393-398.
- Basit S. Vitamin D in health and disease: a literature review. Br J Biomed Sci. 2013; 70: 161-172.
- Hunter D, De Lange M, Snieder H, MacGregor AJ, Swaminathan R, Thakker RV, et al. Genetic contribution to bone metabolism, calcium excretion, and vitamin D and parathyroid hormone regulation. J Bone Miner Res. 2001; 16: 371-378.
- Shea MK, Benjamin EJ, Dupuis J, Massaro JM, Jacques PF, D'Agostino RB Sr, et al. Genetic and non-genetic correlates of vitamins K and D. Eur J Clin Nutr. 2009; 63: 458-464.
- Wjst M, Altmüller J, Braig C, Bahnweg M, André E. A genome-wide linkage scan for 25-OH-D(3) and 1,25-(OH)2-D3 serum levels in asthma families. J Steroid Biochem Mol Biol. 2007; 103: 799-802.
- Omdahl JL, Bobrovnikova EV, Annalora A, Chen P, Serda R. Expression, structure-function, and molecular modeling of vitamin D P450s. J Cell Biochem. 2003; 88: 356-362.
- Gocek E, Studzinski GP. Vitamin D and differentiation in cancer. Crit Rev Clin Lab Sci. 2009; 46: 190-209.
- Simboli-Campbell M, Narvaez CJ, van Weelden K, Tenniswood M, Welsh J. Comparative effects of 1,25(OH)2D3 and EB1089 on cell cycle kinetics and apoptosis in MCF-7 breast cancer cells. Breast Cancer Res Treat. 1997; 42: 31-41.
- Flores O, Wang Z, Knudsen KE, Burnstein KL. Nuclear Targeting of Cyclin-Dependent Kinase 2 Reveals Essential Roles of Cyclin-Dependent Kinase 2 Localization and Cyclin E in Vitamin D-Mediated Growth Inhibition. Endocrinology. 2010; 151: 896-908.
- Jensen SS, Madsen MW, Lukas J, Binderup L, Bartek J. Inhibitory effects of 1alpha,25-dihydroxyvitamin D(3) on the G(1)-S phase-controlling machinery. Mol Endocrinol. 2001; 15: 1370-1380.
- Liu M, Lee MH, Cohen M, Bommakanti M, Freedman LP. Transcriptional Activation of the Cdk Inhibitor p21 by Vitamin D3 Leads to the Induced Differentiation of the Myelomonocytic Cell Line U937. Genes Dev. 1996; 10: 142-153.
- Rohan JN, Weigel NL. 1Alpha,25-dihydroxyvitamin D3 reduces c-Myc expression, inhibiting proliferation and causing G1 accumulation in C4-2 prostate cancer cells. Endocrinology. 2009; 150: 2046-2054.
- Blutt SE, McDonnell TJ, Polek TC, Weigel NL. Calcitriol-induced apoptosis in LNCaP cells is blocked by overexpression of Bcl-2. Endocrinology. 2000; 141: 10-17.
- Lamprecht SA, Lipkin M. Chemoprevention of colon cancer by calcium, vitamin D and folate: molecular mechanisms. Nat Rev Cancer. 2003; 3: 601-614.
- Wu S, Liao AP, Xia Y, Li YC, Li JD, Sartor RB, et al. Vitamin D receptor negatively regulates bacterial-stimulated NF-kappaB activity in intestine. Am J Pathol. 2010; 177: 686-697.
- Cheung FS, Lovicu FJ, Reichardt JK. Current progress in using vitamin D and its analogs for cancer prevention and treatment. Expert Rev Anticancer Ther. 2012; 12: 811-837.
- Krishnan AV, Feldman D. Regulation of Vitamin D Receptor Abundance. Academic Press. 1997; 179-200.
- Krishnan AV, Trump DL, Johnson CS, Feldman D. The role of vitamin D in cancer prevention and treatment. Endocrinol Metab Clin North Am. 2010; 39: 401-418, table of contents.
- Leyssens C, Verlinden L, Verstuyf A. Antineoplastic effects of 1,25(OH)2D3 and its analogs in breast, prostate and colorectal cancer. Endocr Relat Cancer. 2013; 20: R31-47.
- Mehta RG, Peng X, Alimirah F, Murillo G, Mehta R. Vitamin D and breast cancer: emerging concepts. Cancer Lett. 2013; 334: 95-100.
- Pereira F, Larriba MJ, Muñoz A. Vitamin D and colon cancer. Endocr Relat Cancer. 2012; 19: R51-71.
- Tang JY, Fu T, Lau C, Oh DH, Bikle DD, Asgari MM. Vitamin D in cutaneous carcinogenesis: part I. J Am Acad Dermatol. 2012; 67: 803.
- Thorne J, Campbell MJ. The vitamin D receptor in cancer. Proc Nutr Soc. 2008; 67: 115-127.
- Trump DL, Deeb KK, Johnson CS. Vitamin D: considerations in the continued development as an agent for cancer prevention and therapy. Cancer J. 2010; 16: 1-9.
- Deeb KK, Trump DL, Johnson CS. Vitamin D signalling pathways in cancer: potential for anticancer therapeutics. Nat Rev Cancer. 2007; 7: 684-700.
- Fleet JC. Molecular actions of vitamin D contributing to cancer prevention. Mol Aspects Med. 2008; 29: 388-396.
- Fleet JC1, DeSmet M, Johnson R, Li Y. Vitamin D and cancer: a review of molecular mechanisms. Biochem J. 2012; 441: 61-76.
- Haussler MR, Whitfield GK, Kaneko I, Haussler CA, Hsieh D, Hsieh JC, et al. Molecular mechanisms of vitamin D action. Calcif Tissue Int. 2013; 92: 77-98.
- Swami S, Krishnan AV, Wang JY, Jensen K, Peng L, Albertelli MA, et al. Inhibitory effects of calcitriol on the growth of MCF-7 breast cancer xenografts in nude mice: selective modulation of aromatase expression in vivo. Horm Cancer. 2011; 2: 190-202.
- Cohen-Lahav M, Shany S, Tobvin D, Chaimovitz C, Douvdevani A. Vitamin D decreases NFkappaB activity by increasing IkappaBalpha levels. Nephrol Dial Transplant. 2006; 21: 889-897.
- Yu XP, Bellido T, Manolagas SC. Down-regulation of NF-kappa B protein levels in activated human lymphocytes by 1,25-dihydroxyvitamin D3. Proc Natl Acad Sci U S A. 1995; 92: 10990-10994.
- Ananthakrishnan AN, Khalili H, Higuchi LM, Bao Y, Korzenik JR, Giovannucci EL, et al. Higher predicted vitamin D status is associated with reduced risk of Crohn's disease. Gastroenterology. 2012; 142: 482-489.
- Cantorna MT. Vitamin D and its role in immunology: multiple sclerosis, and inflammatory bowel disease. Prog Biophys Mol Biol. 2006; 92: 60-64.
- Froicu M, Weaver V, Wynn TA, McDowell MA, Welsh JE, Cantorna MT. A crucial role for the vitamin D receptor in experimental inflammatory bowel diseases. Mol Endocrinol. 2003; 17: 2386-2392.
- Cantorna MT, Mahon BD. Mounting evidence for vitamin D as an environmental factor affecting autoimmune disease prevalence. Exp Biol Med (Maywood). 2004; 229: 1136-1142.
- Wang TT, Dabbas B, Laperriere D, Bitton AJ, Soualhine H, Tavera-Mendoza LE, et al. Direct and indirect induction by 1,25-dihydroxyvitamin D3 of the NOD2/CARD15-defensin beta2 innate immune pathway defective in Crohn disease. J Biol Chem. 2010; 285: 2227-2231.
- Vaidya A, Forman JP, Hopkins PN, Seely EW, Williams JS. 25-Hydroxyvitamin D is Associated with Plasma Renin Activity and the Pressor Response to Dietary Sodium Intake in Caucasians. J. Renin Angiotensin Aldosterone Syst. 2011; 12: 311-319.
- Vimaleswaran KS, Cavadino A, Berry DJ; LifeLines Cohort Study investigators, Jorde R, Dieffenbach AK, Lu C, et al. Association of vitamin D status with arterial blood pressure and hypertension risk: a mendelian randomisation study. Lancet Diabetes Endocrinol. 2014; 2: 719-729.
- Wang L, Chu A, Buring JE, Ridker PM, Chasman DI, Sesso HD. Common genetic variations in the vitamin D pathway in relation to blood pressure. Am J Hypertens. 2014; 27: 1387-1395.
- Cottone S, Guarino L, Arsena R, Scazzone C, Tornese F, Guarneri M, et al. Vitamin D receptor gene polymorphisms and plasma renin activity in essential hypertensive individuals. J Hum Hypertens. 2015; 29: 483-487.
- Ndiaye NC, Said el S, Stathopoulou MG, Siest G, Tsai MY, Visvikis-Siest S. Epistatic study reveals two genetic interactions in blood pressure regulation. BMC Med Genet. 2013; 14: 2.
- Testa A, Mallamaci F, Benedetto FA, Pisano A, Tripepi G, Malatino L, et al. Vitamin D receptor (VDR) gene polymorphism is associated with left ventricular (LV) mass and predicts left ventricular hypertrophy (LVH) progression in end-stage renal disease (ESRD) patients. J Bone Miner Res. 2010; 25: 313-319.
- Santoro D, Caccamo D, Gagliostro G, Ientile R, Benvenga S, Bellinghieri G, et al. Vitamin D metabolism and activity as well as genetic variants of the vitamin D receptor (VDR) in chronic kidney disease patients. J Nephrol. 2013; 26: 636-644.
- El-Shehaby AM, El-Khatib MM, Marzouk S, Battah AA. Relationship of BsmI polymorphism of vitamin D receptor gene with left ventricular hypertrophy and atherosclerosis in hemodialysis patients. Scand J Clin Lab Invest. 2013; 73: 75-81.
- Santoro D, Gagliostro G, Alibrandi A, Ientile R, Bellinghieri G, Savica V, et al. Vitamin D receptor gene polymorphism and left ventricular hypertrophy in chronic kidney disease. Nutrients. 2014; 6: 1029-1037.
- Santoro D, Buemi M, Gagliostro G, Vecchio M, Currò M, Ientile R, et al. Association of VDR gene polymorphisms with heart disease in chronic kidney disease patients. Clin Biochem. 2015; .
- Cooper JD, Smyth DJ, Walker NM, Stevens H, Burren OS, Wallace C, et al. Inherited Variation in Vitamin D Genes is Associated with Predisposition to Autoimmune Disease Type 1 Diabetes. Diabetes. 2011; 60: 1624-1631.
- Tizaoui K, Kaabachi W, Hamzaoui A, Hamzaoui K. Contribution of VDR polymorphisms to type 1 diabetes susceptibility: Systematic review of case-control studies and meta-analysis. J Steroid Biochem Mol Biol. 2014; 143: 240-249.
- Wang Q, Xi B, Reilly KH, Liu M, Fu M. Quantitative assessment of the associations between four polymorphisms (FokI, ApaI, BsmI, TaqI) of vitamin D receptor gene and risk of diabetes mellitus. Mol Biol Rep. 2012; 39: 9405-9414.
- Simmons JD, Mullighan C, Welsh KI, Jewell DP. Vitamin D receptor gene polymorphism: association with Crohn's disease susceptibility. Gut. 2000; 47: 211-214.
- Jurutka PW, Bartik L, Whitfield GK, Mathern DR, Barthel TK, Gurevich M, et al. Vitamin D receptor: key roles in bone mineral pathophysiology, molecular mechanism of action, and novel nutritional ligands. J Bone Miner Res. 2007; 22 Suppl 2: V2-10.
- Mitra S, Desai M, Ikram Khatkhatay M. Vitamin D receptor gene polymorphisms and bone mineral density in postmenopausal Indian women. Maturitas. 2006; 55: 27-35.
- Ferrari SL. Osteoporosis, vitamin D receptor gene polymorphisms and response to diet. World Rev Nutr Diet. 2001; 89: 83-92.
- Ferrari S, Bonjour JP, Rizzoli R. The vitamin d receptor gene and calcium metabolism. Trends Endocrinol Metab. 1998; 9: 259-265.
- Gray RW, Omdahl JL, Ghazarian JG, DeLuca HF. 25-Hydroxycholecalciferol-1-hydroxylase. Subcellular location and properties. J Biol Chem. 1972; 247: 7528-7532.
- Wu S, Sun J. Vitamin D, vitamin D receptor, and macroautophagy in inflammation and infection. Discov Med. 2011; 11: 325-335.
- Ramagopalan SV, Heger A, Berlanga AJ, Maugeri NJ, Lincoln MR, Burrell A, et al. A ChIP-seq defined genome-wide map of vitamin D receptor binding: associations with disease and evolution. Genome Res. 2010; 20: 1352-1360.
- Kang SW, Kim SH, Lee N, Lee WW, Hwang KA, Shin MS, et al. 1,25-Dihyroxyvitamin D3 promotes FOXP3 expression via binding to vitamin D response elements in its conserved noncoding sequence region. J Immunol. 2012; 188: 5276-5282.
- Wu S, Zhang YG, Lu R, Xia Y, Zhou D, Petrof EO, et al. Intestinal epithelial vitamin D receptor deletion leads to defective autophagy in colitis. Gut. 2015; 64: 1082-1094.
- Gombart AF, Borregaard N, Koeffler HP. Human Cathelicidin Antimicrobial Peptide (CAMP) Gene is a Direct Target of the Vitamin D Receptor and is Strongly Up-Regulated in Myeloid Cells by 1, 25-Dihydroxyvitamin D3. The FASEB journal. 2005; 19: 1067-1077.
- Eloranta JJ, Wenger C, Mwinyi J, Hiller C, Gubler C, Vavricka SR, et al. Association of a Common Vitamin D-Binding Protein Polymorphism with Inflammatory Bowel Disease. Pharmacogenet Genomics. 2011; 21: 559-564.
- Kong J, Zhang Z, Musch MW, Ning G, Sun J, Hart J, et al. Novel role of the vitamin D receptor in maintaining the integrity of the intestinal mucosal barrier. Am J Physiol Gastrointest Liver Physiol. 2008; 294: G208-216.
- Visser R, Barrett K, McCole D. Regulation of Epithelial Barrier Function by Crohn's Disease Associated Gene PTPN2. In BMC Proceedings 2012, 6(Suppl 4):O41, Dublin, Ireland, 4-5 November 2011; pp. O41.
- Godchaux W 3rd, Leadbetter ER. Sulfonolipids of gliding bacteria. Structure of the N-acylaminosulfonates. J Biol Chem. 1984; 259: 2982-2990.
- Marshall TG. Bacterial Capnine Blocks Transcription of Human Antimicrobial Peptides. In Metagenomics. 2007; University of California, San Diego, California, 11 July 2007.
- Salazar JC, Duhnam-Ems S, La Vake C, Cruz AR, Moore MW, Caimano MJ, et al. Activation of Human Monocytes by Live Borrelia Burgdorferi Generates TLR2-Dependent and-Independent Responses which Include Induction of IFN-?. PLoS pathog. 2009; 5: e1000444.
- Xu Y, Xie J, Li Y, Yue J, Chen J, Chunyu L, et al. Using a cDNA microarray to study cellular gene expression altered by Mycobacterium tuberculosis. Chin Med J (Engl). 2003; 116: 1070-1073.
- Roy S, Frodsham A, Saha B, Hazra SK, Mascie-Taylor CG, Hill AV. Association of vitamin D receptor genotype with leprosy type. J Infect Dis. 1999; 179: 187-191.
- He Q, Ananaba GA, Patrickson J, Pitts S, Yi Y, Yan F, et al. Chlamydial infection in vitamin D receptor knockout mice is more intense and prolonged than in wild-type mice. J Steroid Biochem Mol Biol. 2013; 135: 7-14.
- Liu PT, Wheelwright M, Teles R, Komisopoulou E, Edfeldt K, Ferguson B, et al. MicroRNA-21 targets the vitamin D-dependent antimicrobial pathway in leprosy. Nat Med. 2012; 18: 267-273.
- Yenamandra SP, Lundin A, Arulampalam V, Yurchenko M, Pettersson S, Klein G, et al. Expression profile of nuclear receptors upon Epstein -- Barr virus induced B cell transformation. Exp Oncol. 2009; 31: 92-96.
- Cardoso CC, Pereira AC, de Sales Marques C, Moraes MO. Leprosy Susceptibility: Genetic Variations Regulate Innate and Adaptive Immunity, and Disease Outcome. Future Microbiol. 2011; 6: 533-549.
- Sales-Marques C, Salomão H, Fava VM, Alvarado-Arnez LE, Amaral EP, Cardoso CC, et al. NOD2 and CCDC122-LACC1 genes are associated with leprosy susceptibility in Brazilians. Hum Genet. 2014; 133: 1525-1532.
- Alter A, de Léséleuc L, Van Thuc N, Thai VH, Huong NT, Ba NN, et al. Genetic and functional analysis of common MRC1 exon 7 polymorphisms in leprosy susceptibility. Hum Genet. 2010; 127: 337-348.
- Gupta SC, Patchva S, Aggarwal BB. Therapeutic roles of curcumin: lessons learned from clinical trials. AAPS J. 2013; 15: 195-218.
- Aggarwal BB, Gupta SC, Sung B. Curcumin: an orally bioavailable blocker of TNF and other pro-inflammatory biomarkers. Br J Pharmacol. 2013; 169: 1672-1692.
- Batie S, Lee JH, Jama RA, Browder DO, Montano LA, Huynh CC, et al. Synthesis and biological evaluation of halogenated curcumin analogs as potential nuclear receptor selective agonists. Bioorg Med Chem. 2013; 21: 693-702.
- Holt PR, Katz S, Kirshoff R. Curcumin therapy in inflammatory bowel disease: a pilot study. Dig Dis Sci. 2005; 50: 2191-2193.
- Suskind DL, Wahbeh G, Burpee T, Cohen M, Christie D, Weber W. Tolerability of curcumin in pediatric inflammatory bowel disease: a forced-dose titration study. J Pediatr Gastroenterol Nutr. 2013; 56: 277-279.
- Shehzad A, Rehman G, Lee YS. Curcumin in inflammatory diseases. Biofactors. 2013; 39: 69-77.
- Dhawan P, Christakos S. Novel regulation of 25-hydroxyvitamin D3 24-hydroxylase (24(OH)ase) transcription by glucocorticoids: cooperative effects of the glucocorticoid receptor, C/EBP beta, and the Vitamin D receptor in 24(OH)ase transcription. J Cell Biochem. 2010; 110: 1314-1323.
- Miller AV, Ranatunga SK. Immunotherapies in rheumatologic disorders. Med Clin North Am. 2012; 96: 475-496, ix-x.
- Smirnoff P, Liel Y, Gnainsky J, Shany S, Schwartz B. The Protective Effect of Estrogen Against Chemically Induced Murine Colon Carcinogenesis is Associated with Decreased CpG Island Methylation and Increased mRNA and Protein Expression of the Colonic Vitamin D Receptor. Oncol. Res. 1999; 11: 255-264.
- Chen TL, Feldman D. Retinoic acid modulation of 1,25(OH)2 vitamin D3 receptors and bioresponse in bone cells: species differences between rat and mouse. Biochem Biophys Res Commun. 1985; 132: 74-80.
- McGrath JJ, Saha S, Burne TH, Eyles DW. A systematic review of the association between common single nucleotide polymorphisms and 25-hydroxyvitamin D concentrations. J Steroid Biochem Mol Biol. 2010; 121: 471-477.
- Köstner K, Denzer N, Müller CS, Klein R, Tilgen W, Reichrath J. The relevance of vitamin D receptor (VDR) gene polymorphisms for cancer: a review of the literature. Anticancer Res. 2009; 29: 3511-3536.
- Pouresmaeili F, Jamshidi J, Azargashb E, Samangouee S. Association between Vitamin D Receptor Gene BsmI Polymorphism and Bone Mineral Density in A Population of 146 Iranian Women. Cell J. 2013; 15: 75-82.
- Lambrinoudaki I, Kaparos G, Armeni E, Alexandrou A, Damaskos C, Logothetis E, et al. BsmI vitamin D receptor's polymorphism and bone mineral density in men and premenopausal women on long-term antiepileptic therapy. Eur J Neurol. 2011; 18: 93-98.
- Horst-Sikorska W, Dytfeld J, Wawrzyniak A, Marcinkowska M, Michalak M, Franek E, et al. Vitamin D receptor gene polymorphisms, bone mineral density and fractures in postmenopausal women with osteoporosis. Mol Biol Rep. 2013; 40: 383-390.
- Mohammadi Z, Fayyazbakhsh F, Ebrahimi M, Amoli MM, Khashayar P, Dini M, et al. Association between vitamin D receptor gene polymorphisms (Fok1 and Bsm1) and osteoporosis: a systematic review. J Diabetes Metab Disord. 2014; 13: 98.
- Cooper GS, Umbach DM. Are vitamin D receptor polymorphisms associated with bone mineral density? A meta-analysis. J Bone Miner Res. 1996; 11: 1841-1849.
- Michos ED, Melamed ML. Vitamin D and cardiovascular disease risk. Curr Opin Clin Nutr Metab Care. 2008; 11: 7-12.
- Ortlepp JR, Hoffmann R, Ohme F, Lauscher J, Bleckmann F, Hanrath P. The vitamin D receptor genotype predisposes to the development of calcific aortic valve stenosis. Heart. 2001; 85: 635-638.
- Kammerer CM, Dualan AA, Samollow PB, Périssé AR, Bauer RL, MacCluer JW, et al. Bone mineral density, carotid artery intimal medial thickness, and the vitamin D receptor BsmI polymorphism in Mexican American women. Calcif Tissue Int. 2004; 75: 292-298.
- Van Schooten FJ, Hirvonen A, Maas LM, De Mol BA, Kleinjans JC, Bell DA, et al. Putative Susceptibility Markers of Coronary Artery Disease: Association between VDR Genotype, Smoking, and Aromatic DNA Adduct Levels in Human Right Atrial Tissue. FASEB J. 1998; 12: 1409-1417.
- Ortlepp JR, von Korff A, Hanrath P, Zerres K, Hoffmann R. Vitamin D receptor gene polymorphism BsmI is not associated with the prevalence and severity of CAD in a large-scale angiographic cohort of 3441 patients. Eur J Clin Invest. 2003; 33: 106-109.
- Hossein-Nezhad A, Eshaghi SM, Maghbooli Z, Mirzaei K, Shirzad M, Curletto B, et al. The role of vitamin D deficiency and vitamin d receptor genotypes on the degree of collateralization in patients with suspected coronary artery disease. Biomed Res Int. 2014; 2014: 304250.
- Qin WH, Wang HX, Qiu JL, Huang XB, Huang Y, Wu NR, et al. A meta-analysis of association of vitamin D receptor BsmI gene polymorphism with the risk of type 1 diabetes mellitus. J Recept Signal Transduct Res. 2014; 34: 372-377.
- Wang G, Zhang Q, Xu N, Xu K, Wang J, He W, et al. Associations between two polymorphisms (FokI and BsmI) of vitamin D receptor gene and type 1 diabetes mellitus in Asian population: a meta-analysis. PLoS One. 2014; 9: e89325.
- Li L, Wu B, Liu JY, Yang LB. Vitamin D receptor gene polymorphisms and type 2 diabetes: a meta-analysis. Arch Med Res. 2013; 44: 235-241.
- Ferrarezi D, Bellili-Muñoz N, Dubois-Laforgue D, Cheurfa N, Lamri A, Reis A, et al. Allelic Variations of the Vitamin D Receptor (VDR) Gene are Associated with Increased Risk of Coronary Artery Disease in Type 2 Diabetics: The DIABHYCAR Prospective Study. Diabetes Metab. 2013; 39: 263-270.
- Xue LN, Xu KQ, Zhang W, Wang Q, Wu J, Wang XY. Associations between vitamin D receptor polymorphisms and susceptibility to ulcerative colitis and Crohn's disease: a meta-analysis. Inflamm Bowel Dis. 2013; 19: 54-60.
- Bentley RW, Keown D, Merriman TR, Raj Krishnan M, Gearry RB, Barclay ML, et al. Vitamin D receptor gene polymorphism associated with inflammatory bowel disease in New Zealand males. Aliment Pharmacol Ther. 2011; 33: 855-856.
- Wang L, Wang ZT, Hu JJ, Fan R, Zhou J, Zhong J. Polymorphisms of the vitamin D receptor gene and the risk of inflammatory bowel disease: a meta-analysis. Genet Mol Res. 2014; 13: 2598-2610.
- Carvalho AY, Bishop KS, Han DY, Ellett S, Jesuthasan A, Lam WJ, et al. The role of Vitamin D level and related single nucleotide polymorphisms in Crohn's disease. Nutrients. 2013; 5: 3898-3909.
- Junker K, Ficarra V, Kwon ED, Leibovich BC, Thompson RH, Oosterwijk E. Potential role of genetic markers in the management of kidney cancer. Eur Urol. 2013; 63: 333-340.
- Kwon YJ, Lee SJ, Koh JS, Kim SH, Lee HW, Kang MC, et al. Genome-wide analysis of DNA methylation and the gene expression change in lung cancer. J Thorac Oncol. 2012; 7: 20-33.
- Lokk K, Vooder T, Kolde R, Välk K, Võsa U, Roosipuu R, et al. Methylation markers of early-stage non-small cell lung cancer. PLoS One. 2012; 7: e39813.
- Peters I, Eggers H, Atschekzei F, Hennenlotter J, Waalkes S, Tränkenschuh W, et al. GATA5 CpG island methylation in renal cell cancer: a potential biomarker for metastasis and disease progression. BJU Int. 2012; 110: E144-152.
- Levin GP, Robinson-Cohen C, de Boer IH, Houston DK, Lohman K, Liu Y, et al. Genetic variants and associations of 25-hydroxyvitamin D concentrations with major clinical outcomes. JAMA. 2012; 308: 1898-1905.
- Bouillon R, Carmeliet G, Verlinden L, van Etten E, Verstuyf A, Luderer HF, et al. Vitamin D and human health: lessons from vitamin D receptor null mice. Endocr Rev. 2008; 29: 726-776.
- Wang TJ, Zhang F, Richards JB, Kestenbaum B, van Meurs JB, Berry D, et al. Common genetic determinants of vitamin D insufficiency: a genome-wide association study. Lancet. 2010; 376: 180-188.
- Janssens W, Bouillon R, Claes B, Carremans C, Lehouck A, Buysschaert I, et al. Vitamin D deficiency is highly prevalent in COPD and correlates with variants in the vitamin D-binding gene. Thorax. 2010; 65: 215-220.
- Wang G, Li Y, Li L, Yu F, Cui L, Ba Y, et al. Association of the vitamin D binding protein polymorphisms with the risk of type 2 diabetes mellitus: a meta-analysis. BMJ Open. 2014; 4: e005617.
- Rebhan M, Chalifa-Caspi V, Prilusky J, Lancet D. GeneCards: a novel functional genomics compendium with automated data mining and query reformulation support. Bioinformatics. 1998; 14: 656-664.
- Cooke NE, Haddad JG. Vitamin D binding protein (Gc-globulin). Endocr Rev. 1989; 10: 294-307.
- Lauridsen AL, Vestergaard P, Nexo E. Mean serum concentration of vitamin D-binding protein (Gc globulin) is related to the Gc phenotype in women. Clin Chem. 2001; 47: 753-756.
- Dahl B, Schiødt FV, Ott P, Wians F, Lee WM, Balko J, et al. Plasma concentration of Gc-globulin is associated with organ dysfunction and sepsis after injury. Crit Care Med. 2003; 31: 152-156.
- Constans J, Arlet P, Viau M, Bouissou C. Unusual sialilation of the serum DBP associated with the Gc 1 allele in alcoholic cirrhosis of the liver. Clin Chim Acta. 1983; 130: 219-230.
- Kamboh MI, Ferrell RE. Ethnic variation in vitamin D-binding protein (GC): a review of isoelectric focusing studies in human populations. Hum Genet. 1986; 72: 281-293.
- Speeckaert M, Huang G, Delanghe JR, Taes YE. Biological and Clinical Aspects of the Vitamin D Binding Protein (Gc-Globulin) and its Polymorphism. Clinica chimica acta. 2006; 372: 33-42.
- Wilson RT, Bortner JD, Roff A, Das A, Battaglioli EJ, Richie JP, et al. Genetic and Environmental Influences on Plasma Vitamin D Binding Protein Concentrations. Transl Res. 2015; 165: 667-676.
- Braun A, Brandhofer A, Cleve H. Interaction of the Vitamin D-binding Protein (Groupspecific Component) and its Ligand 25-hydroxyvitamin D3: Binding Differences of the various Genetic Types Disclosed by Isoelectric Focusing. Electrophoresis. 1990; 11: 478-483.
- Rejnmark L, Lauridsen AL, Vestergaard P, Heickendorff L, Andreasen F, Mosekilde L. Diurnal rhythm of plasma 1,25-dihydroxyvitamin D and vitamin D-binding protein in postmenopausal women: relationship to plasma parathyroid hormone and calcium and phosphate metabolism. Eur J Endocrinol. 2002; 146: 635-642.
- Signorello LB, Shi J, Cai Q, Zheng W, Williams SM, Long J, et al. Common variation in vitamin D pathway genes predicts circulating 25-hydroxyvitamin D Levels among African Americans. PLoS One. 2011; 6: e28623.
- Ahn J, Yu K, Stolzenberg-Solomon R, Simon KC, McCullough ML, Gallicchio L, et al. Genome-wide association study of circulating vitamin D levels. Hum Mol Genet. 2010; 19: 2739-2745.
- Lu L, Sheng H, Li H, Gan W, Liu C, Zhu J, et al. Associations between common variants in GC and DHCR7/NADSYN1 and vitamin D concentration in Chinese Hans. Hum Genet. 2012; 131: 505-512.
- Hibler EA, Hu C, Jurutka PW, Martinez ME, Jacobs ET. Polymorphic Variation in the GC and CASR Genes and Associations with Vitamin D Metabolite Concentration and Metachronous Colorectal Neoplasia. Epidemiol Biomarkers Prev. 2012; 21: 368-375.
- Sinotte M, Diorio C, Bérubé S, Pollak M, Brisson J. Genetic polymorphisms of the vitamin D binding protein and plasma concentrations of 25-hydroxyvitamin D in premenopausal women. Am J Clin Nutr. 2009; 89: 634-640.
- Miller WL. Steroid hormone synthesis in mitochondria. Mol Cell Endocrinol. 2013; 379: 62-73.
- Chen TC, Chimeh F, Lu Z, Mathieu J, Person KS, Zhang A, et al. Factors that influence the cutaneous synthesis and dietary sources of vitamin D. Arch Biochem Biophys. 2007; 460: 213-217.
- Kuan V, Martineau AR, Griffiths CJ, Hyppönen E, Walton R. DHCR7 mutations linked to higher vitamin D status allowed early human migration to northern latitudes. BMC Evol Biol. 2013; 13: 144.
- Cortes VA, Busso D, Mardones P, Maiz A, Arteaga A, Nervi F, et al. Advances in the physiological and pathological implications of cholesterol. Biol Rev Camb Philos Soc. 2013; 88: 825-843.
- Nolan-Clark D, Tapsell LC, Hu R, Han DY, Ferguson LR. Effects of dairy products on crohn's disease symptoms are influenced by fat content and disease location but not lactose content or disease activity status in a New Zealand population. J Am Diet Assoc. 2011; 111: 1165-1172.
- Steffen KM, Cooper ME, Shi M, Caprau D, Simhan HN, Dagle JM, et al. Maternal and fetal variation in genes of cholesterol metabolism is associated with preterm delivery. J Perinatol. 2007; 27: 672-680.
- Swanson JM, Entringer S, Buss C, Wadhwa PD. Developmental origins of health and disease: environmental exposures. Semin Reprod Med. 2009; 27: 391-402.
- Witsch-Baumgartner M, Sawyer H, Haas D. Clinical utility gene card for: Smith-Lemli-Opitz Syndrome [SLOS]. Eur J Hum Genet. 2013; 21.
- SMITH DW, LEMLI L, OPITZ JM. A NEWLY RECOGNIZED SYNDROME OF MULTIPLE CONGENITAL ANOMALIES. J Pediatr. 1964; 64: 210-217.
- Vimaleswaran KS, Cavadino A, Hyppönen E. APOA5 genotype influences the association between 25-hydroxyvitamin D and high density lipoprotein cholesterol. Atherosclerosis. 2013; 228: 188-192.
- Shirts BH, Howard MT, Hasstedt SJ, Nanjee MN, Knight S, Carlquist JF, et al. Vitamin D dependent effects of APOA5 polymorphisms on HDL cholesterol. Atherosclerosis. 2012; 222: 167-174.
- Slater NA, Rager ML, Havrda DE, Harralson AF. Genetic Variation in CYP2R1 and GC Genes Associated With Vitamin D Deficiency Status. J Pharm Pract. 2015; .
- Ramos-Lopez E, Brück P, Jansen T, Herwig J, Badenhoop K. CYP2R1 (vitamin D 25-hydroxylase) gene is associated with susceptibility to type 1 diabetes and vitamin D levels in Germans. Diabetes Metab Res Rev. 2007; 23: 631-636.
- Bu FX, Armas L, Lappe J, Zhou Y, Gao G, Wang HW, et al. Comprehensive association analysis of nine candidate genes with serum 25-hydroxy vitamin D levels among healthy Caucasian subjects. Hum Genet. 2010; 128: 549-556.
- Thacher TD, Fischer PR, Singh RJ, Roizen J, Levine MA. CYP2R1 Mutations Impair Generation of 25-hydroxyvitamin D and Cause an Atypical Form of Vitamin D Deficiency. J Clin Endocrinol Metab. 2015; 100: E1005-1013.
- Al Mutair AN, Nasrat GH, Russell DW. Mutation of the CYP2R1 vitamin D 25-hydroxylase in a Saudi Arabian family with severe vitamin D deficiency. J Clin Endocrinol Metab. 2012; 97: E2022-2025.
- Hollis BW. Assessment of vitamin D status and definition of a normal circulating range of 25-hydroxyvitamin D. Curr Opin Endocrinol Diabetes Obes. 2008; 15: 489-494.
- Fraser WD, Milan AM. Vitamin D assays: past and present debates, difficulties, and developments. Calcif Tissue Int. 2013; 92: 118-127.
- Nissen J, Vogel U, Ravn-Haren G, Andersen EW, Madsen KH, Nexo BA, et al. Common Variants in CYP2R1 and GC Genes are both Determinants of Serum 25-Hydroxyvitamin D Concentrations After UVB Irradiation and After Consumption of Vitamin D(3)-Fortified Bread and Milk during Winter in Denmark. Am. J. Clin. Nutr. 2015; 101: 218-227.
- Wjst M, Dold S. Genes, factor X, and allergens: what causes allergic diseases? Allergy. 1999; 54: 757-759.
- Wjst M. The triple T allergy hypothesis. Clin Dev Immunol. 2004; 11: 175.
- Milner JD, Stein DM, McCarter R, Moon RY. Early infant multivitamin supplementation is associated with increased risk for food allergy and asthma. Pediatrics. 2004; 114: 27-32.
- Adorini L, Penna G, Giarratana N, Uskokovic M. Tolerogenic Dendritic Cells Induced by Vitamin D Receptor Ligands Enhance Regulatory T Cells Inhibiting Allograft Rejection and Autoimmune Diseases. J. Cell. Biochem. 2003; 88: 227-233.
- Ewens WJ, Spielman RS. What is the significance of a significant TDT? Hum Hered. 2005; 60: 206-210.
- Wjst M, Altmüller J, Faus-Kessler T, Braig C, Bahnweg M, André E. Asthma families show transmission disequilibrium of gene variants in the vitamin D metabolism and signalling pathway. Respir Res. 2006; 7: 60.
- Barry E.L, Rees JR, Peacock JL, Mott LA, Amos CI, Bostick RM, et al. Genetic Variants in CYP2R1, CYP24A1, and VDR Modify the Efficacy of Vitamin D3 Supplementation for Increasing Serum 25-Hydroxyvitamin D Levels in a Randomized Controlled Trial. J Clin Endocrinol Metab. 2004; 99: E2133-E2137.
- Sollid ST, Hutchinson MY, Fuskevag OM, Joakimsen RM, Jorde R. Large Individual Differences in Serum 25-Hydroxyvitamin D Response to Vitamin D Supplementation: Effects of Genetic Factors, Body Mass Index, and Baseline Concentration. Results from a Randomized Controlled Trial. Horm. Metab. Res. 2015.
- Waterhouse M, Tran B, Armstrong BK, Baxter C, Ebeling PR, English DR, et al. Environmental, personal, and genetic determinants of response to vitamin D supplementation in older adults. J Clin Endocrinol Metab. 2014; 99: E1332-1340.
- Armbrecht HJ, Hodam TL, Boltz MA, Partridge NC, Brown AJ, Kumar VB. Induction of the vitamin D 24-hydroxylase (CYP24) by 1,25-dihydroxyvitamin D3 is regulated by parathyroid hormone in UMR106 osteoblastic cells. Endocrinology. 1998; 139: 3375-3381.
- Jones G, Prosser DE, Kaufmann M. 25-Hydroxyvitamin D-24-hydroxylase (CYP24A1): its important role in the degradation of vitamin D. Arch Biochem Biophys. 2012; 523: 9-18.
- Perwad F, Zhang MY, Tenenhouse HS, Portale AA. Fibroblast growth factor 23 impairs phosphorus and vitamin D metabolism in vivo and suppresses 25-hydroxyvitamin D-1alpha-hydroxylase expression in vitro. Am J Physiol Renal Physiol. 2007; 293: F1577-1583.
- Roff A, Wilson RT. A novel SNP in a vitamin D response element of the CYP24A1 promoter reduces protein binding, transactivation, and gene expression. J Steroid Biochem Mol Biol. 2008; 112: 47-54.
- Schlingmann KP, Kaufmann M, Weber S, Irwin A, Goos C, John U, et al. Mutations in CYP24A1 and idiopathic infantile hypercalcemia. N Engl J Med. 2011; 365: 410-421.
- Jacobs ET, Van Pelt C, Forster RE, Zaidi W, Hibler EA, Galligan MA, et al. CYP24A1 and CYP27B1 polymorphisms modulate vitamin D metabolism in colon cancer cells. Cancer Res. 2013; 73: 2563-2573.
- Babiker AM, Al Gadi I, Al-Jurayyan NA, Al Nemri AM, Al Haboob AA, Al Boukai AA, et al. A novel pathogenic mutation of the CYP27B1 gene in a patient with vitamin D-dependent rickets type 1: a case report. BMC Res Notes. 2014; 7: 783.
- Cooper DN, Ball EV, Stenson PD, Phillips AD, Howells K, Heywood S, et al. The Human Gene Data Base. 2015; 2015.
- Orton SM, Morris AP, Herrera BM, Ramagopalan SV, Lincoln MR, Chao MJ, et al. Evidence for genetic regulation of vitamin D status in twins with multiple sclerosis. Am J Clin Nutr. 2008; 88: 441-447.
- Bahlo M, Booth DR, Broadley SA, Brown MA, Foote SJ, Griffiths LR, et al. Genome-Wide Association Study Identifies New Multiple Sclerosis Susceptibility Loci on Chromosomes 12 and 20. Nat. Genet. 2009; 41: 824-828.
- Cortes A, Field J, Glazov EA, Hadler J, ANZgene Consortium, Stankovich J, et al. Resequencing and Fine-Mapping of the Chromosome 12q13-14 Locus Associated with Multiple Sclerosis Refines the Number of Implicated Genes. Hum. Mol. Genet. 2013; 22: 2283-2292.
- Handel AE, Handunnetthi L, Berlanga AJ, Watson CT, Morahan JM, Ramagopalan SV. The effect of single nucleotide polymorphisms from genome wide association studies in multiple sclerosis on gene expression. PLoS One. 2010; 5: e10142.
- Ross JP, Bernales CQ, Lee JD, Sadovnick AD, Traboulsee AL, Vilariño-Güell C. Analysis of CYP27B1 in multiple sclerosis. J Neuroimmunol. 2014; 266: 64-66.
- Bailey R, Cooper JD, Zeitels L, Smyth DJ, Yang JH, Walker NM, et al. Association of the vitamin D metabolism gene CYP27B1 with type 1 diabetes. Diabetes. 2007; 56: 2616-2621.
- Ramos-Lopez E, Kahles H, Weber S, Kukic A, Penna-Martinez M, Badenhoop K, et al. Gestational diabetes mellitus and vitamin D deficiency: genetic contribution of CYP27B1 and CYP2R1 polymorphisms. Diabetes Obes Metab. 2008; 10: 683-685.
- Zhuang JC, Huang ZY, Zhao GX, Yu H, Li ZX, Wu ZY. Variants of CYP27B1 are associated with both multiple sclerosis and neuromyelitis optica patients in Han Chinese population. Gene. 2015; 557: 236-239.
- Wu GD, Lewis JD, Hoffmann C, Chen YY, Knight R, Bittinger K, et al. Sampling and pyrosequencing methods for characterizing bacterial communities in the human gut using 16S sequence tags. BMC Microbiol. 2010; 10: 206.
- Proctor LM. The Human Microbiome Project in 2011 and beyond. Cell Host Microbe. 2011; 10: 287-291.
- Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A. 2011; 108 Suppl 1: 4554-4561.
- Abubucker S, Segata N, Goll J, Schubert AM, Izard J, Cantarel BL, et al. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput Biol. 2012; 8: e1002358.
- Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006; 124: 837-848.
- Christie B. [More than Human] Buddy can You Spare a Gene. Sci Am. 2012; 306: 36-43.
- Balter M. Taking stock of the human microbiome and disease. Science. 2012; 336: 1246-1247.
- Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013; 504: 446-450.
- Hooper LV, Stappenbeck TS, Hong CV, Gordon JI. Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nat Immunol. 2003; 4: 269-273.
- Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004; 118: 229-241.
- Mantovani A, Cassatella MA, Costantini C, Jaillon S. Neutrophils in the activation and regulation of innate and adaptive immunity. Nat Rev Immunol. 2011; 11: 519-531.
- Joyce SA, Gahan CG. The gut microbiota and the metabolic health of the host. Curr Opin Gastroenterol. 2014; 30: 120-127.
- Duseja A, Chawla YK. Obesity and NAFLD: the role of bacteria and microbiota. Clin Liver Dis. 2014; 18: 59-71.
- Walker AW, Sanderson JD, Churcher C, Parkes GC, Hudspith BN, Rayment N, et al. High-Throughput Clone Library Analysis of the Mucosa-Associated Microbiota Reveals Dysbiosis and Differences between Inflamed and Non-Inflamed Regions of the Intestine in Inflammatory Bowel Disease. BMC Microbiol. 2011; 11: 7-2180-11-7.
- Tamboli CP, Neut C, Desreumaux P, Colombel JF. Dysbiosis in inflammatory bowel disease. Gut. 2004; 53: 1-4.
- Sobhani I, Tap J, Roudot-Thoraval F, Roperch JP, Letulle S, Langella P, et al. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS One. 2011; 6: e16393.
- Wu N, Yang X, Zhang R, Li J, Xiao X, Hu Y, et al. Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microb Ecol. 2013; 66: 462-470.
- Jin D, Wu S, Zhang YG, Lu R, Xia Y, Dong H, et al. Lack of Vitamin D Receptor Causes Dysbiosis and Changes the Functions of the Murine Intestinal Microbiome. Clin Ther. 2015; 37: 996-1009.
- Chen J, Waddell A, Lin Y, Cantorna M. Dysbiosis Caused by Vitamin D Receptor Deficiency Confers Colonization Resistance to Citrobacter Rodentium through Modulation of Innate Lymphoid Cells. Mucosal Immunol. 2014.
- Lakatos PL, Fischer S, Lakatos L, Gal I, Papp J. Current concept on the pathogenesis of inflammatory bowel disease-crosstalk between genetic and microbial factors: pathogenic bacteria and altered bacterial sensing or changes in mucosal integrity take "toll" ? World J Gastroenterol. 2006; 12: 1829-1841.
- Naser SA, Arce M, Khaja A, Fernandez M, Naser N, Elwasila S, et al. Role of ATG16L, NOD2 and IL23R in Crohn's disease pathogenesis. World J Gastroenterol. 2012; 18: 412-424.
- Homer CR, Richmond AL, Rebert NA, Achkar J, McDonald C. ATG16L1 and NOD2 Interact in an Autophagy-Dependent Antibacterial Pathway Implicated in Crohn's Disease Pathogenesis. Gastroenterology. 2010; 139: 1630-1641.
- Cantorna MT, McDaniel K, Bora S, Chen J, James J. Vitamin D, immune regulation, the microbiota, and inflammatory bowel disease. Exp Biol Med (Maywood). 2014; 239: 1524-1530.
- Schauber J, Dorschner RA, Coda AB, Büchau AS, Liu PT, Kiken D, et al. Injury enhances TLR2 function and antimicrobial peptide expression through a vitamin D-dependent mechanism. J Clin Invest. 2007; 117: 803-811.
- Donovan SM, Wang M, Li M, Friedberg I, Schwartz SL, Chapkin RS. Host-microbe interactions in the neonatal intestine: role of human milk oligosaccharides. Adv Nutr. 2012; 3: 450S-5S.
- Wu S, Tao N, German JB, Grimm R, Lebrilla CB. Development of an annotated library of neutral human milk oligosaccharides. J Proteome Res. 2010; 9: 4138-4151.
- Young VB, Schmidt TM. Antibiotic-associated diarrhea accompanied by large-scale alterations in the composition of the fecal microbiota. J Clin Microbiol. 2004; 42: 1203-1206.