
Research Article
Austin J Nutr Metab. 2021; 8(2): 1103.
Amplification of the Hazelnut-Induced Epigenetic Modulation of LDLR Gene Expression in THLE-2 Human Primary Hepatocytes Compared to HepG2 Hepatocarcinoma Cells
Benassi B¹*, Santangeli S², Bacchetta L³ and Pacchierotti F¹
¹Division of Health Protection Technologies, ENEAItalian National Agency for New Technologies, Energy and Sustainable Economic Development, Italy
²Department of Life and Environmental Sciences, Marche Polytechnic University, Italy
³Division of Biotechnologies and Agroindustry, ENEAItalian National Agency for New Technologies, Energy and Sustainable Economic Development, Italy
*Corresponding author: Benassi B, Division of Health Protection Technologies, ENEA-Casaccia, Via Anguillarese 301, 00123 Rome, Italy
Received: February 06, 2021; Accepted: March 06, 2021; Published: March 13, 2021
Abstract
Dietary supplementation with tree nuts, including hazelnuts, has been associated with reduced cardiovascular disease risk factors, due to improved blood lipid profile. In the attempt to identify the molecular mechanism(s) underlying such beneficial effect, we here characterized the response of the human primary hepatocytes (THLE-2 cells) to the administration of an ethanolic extract of hazelnut (Corylus avellana L., cultivar Tonda Gentile Romana) in terms of regulation of the Low-Density Lipoprotein Receptor (LDLR), a major blood cholesterol carrier that positively correlates with improved blood lipids level. We demonstrated that hazelnut (0.004-0.4 mg/ml) does not alter viability and growth of primary liver cells, but significantly stimulates (P<0.05) the LDLR mRNA and membrane protein expression. We also proved that LDLR increase is driven by an epigenetic-based mechanism, as hazelnut treatments trigger a strong DNA hypo-methylation of two CpG-rich regions in the LDLR gene promoter, mapping at -739/-548 and -221/+174 position. The hazelnutmediated hypo-methylation is much stronger and extensive in THLE-2 than in HepG2 human hepatocarcinoma cells, supporting the significance of the liver context for performing studies on dietary supplementation. As overall, our findings demonstrate that hazelnut triggers an epigenetic-dependent regulation of LDLR expression in human primary liver cells in vitro, thus identifying in the LDLR stimulation a molecular mechanism contributing to the health promoting functions of hazelnuts.
Keywords: Hazelnut; LDLR; Epigenetics; DNA methylation; THLE-2 human primary hepatocytes
Abbreviations
CpG: Cytosine-Guanine Dinucleotide separated by a Phosphodiester Bond; CVD: Cardiovascular Disease; DAC: 5-Aza- 2-deoxycytidine; DMEM: Dulbecco’s Modified Eagle’s Medium; DMSO: Dimethyl Sulfoxide; EDTA: Ethylenediaminetetraacetic Acid; FACS: Fluorescence-Activated Single Cell Sorting; FSC-H: Forward Scatter; GAPDH: Glyceraldehyde-3-Phosphate Dehydrogenase; HepG2: Human Hepatocarcinoma Cell Line; hsa, Homo Sapiens; HZN: Hazelnut; LDL: Low-Density Lipoprotein; LDLR: Low-Density Lipoprotein Receptor; 5-mC: 5-Methyl Cytosine; miR: microRNA; PBS: Phosphate Buffer Saline; PI: Propidium Iodide; SSC-H: Side Scatter; SD: Standard Deviation; THLE-2 cells: Hepatic SV40 Large T-Antigen Immortalized Human Liver Epithelial Cells; TSS: Transcription Starting Site.
Introduction
Several clinical and epidemiological evidences have highlighted the beneficial effects of a dietary supplementation with tree nuts in terms of plasma lipid levels and prevention of Cardiovascular Diseases (CVDs) [1-3], although no clear identification of the molecular pathways underlying such effect has been provided. In this context, we recently reported that an ethanolic extract of hazelnut (Corylus avellana L.) can trigger an epigenetic-dependent stimulation of the expression of the Low-Density Lipoprotein Receptor (LDLR)-a major blood cholesterol carrier- in a model of human hepatocarcinoma cell line (HepG2) [4].
Despite HepG2 is a widely used cell model for mimicking liver metabolism, the use of a tumor cell line may lead to confounding interpretation of liver response to metabolites as -unlike normal hepatocytes- HepG2 displays a limited metabolizing capacity [5,6]. Hence, there is a growing need for alternative-non-cancer models that might better represent liver physiology; at present, primary human hepatocytes represent an important tool for research purposes [7], and the THLE cell line (hepatic SV40 large T-antigen immortalized human liver epithelial cells) has been reported as promising model for investigation of liver metabolism [8,9]. The choice for the proper liver experimental model is of primary importance, especially when epigenetic mechanisms are involved. The liver epigenome is extremely sensitive to the environmental hits and, as such, metabolic risk factors -such as obesity, excessive alcohol consumption and viral hepatitiscan dramatically affect the hepatic epigenome in primary hepatocytes. Specifically, some experimental evidences highlighted the different epigenetic pattern of HepG2 compared to THLE experimental models, in terms of microRNAs (miR) expression, DNA methylation response, as well as histone acetylation levels [10,11]. The control of LDLR expression has been also recently characterized in terms of epigenetic regulation in HepG2 cells and animal models, via either a miR-dependent [12] or a histone modification-mediated pathway [13]. In this context, we demonstrated that the DNA methylation pattern of LDLR gene promoter is modified in HepG2 cells by treatment with a hazelnut extract, and disclosed the first epigenetic pathway in LDLR gene expression control by nut species [4].
Based on these findings, we hypothesize that the molecular pathway controlling LDLR expression in liver cells may differ in primary liver cells compared to hepatocarcinoma cells, and may differently affect the liver cells response to the hazelnut extract administration. Hence, we here aim at characterizing the response to hazelnut in a more physiological liver experimental model, i.e. the THLE-2 primary human hepatocytes, in terms of LDLR expression and underlying molecular mechanisms.
Methods
Chemicals
Phosphate Buffer Saline (PBS), DMEM (Dulbecco’s Modified Eagle’s Medium) culture medium, culture supplements, and trypsin- EDTA (ethylenediaminetetraacetic acid) were obtained from Euroclone (Milan, Italy). LHC-8 culture medium was purchased from Gibco (Thermo Fisher Scientific, Waltham, MA, USA). The fetal bovine serum was obtained from Hyclone (Logan, UT, USA). 5-Aza-2-Deoxycytidine (DAC), bovine serum albumin, collagen, Dimethyl Sulfoxide (DMSO), epidermal growth factor, fibronectin, phosphoethanolamine, and Trypan blue solution (0.4%) were obtained from SIGMA-Aldrich (St Louis, MO, USA). Ethanol was obtained from CARLO ERBA Reagents (Milan, Italy).
Preparation of the hazelnut liquid extract (HZN)
Preparation of the ethanolic extract of hazelnut (Corylus avellana L., cultivar Tonda Gentile Romana, Coopernocciola srl, Vico Matrino, VT, Italy) was carried out as previously detailed [4]. Briefly, 20 grams of raw materials were homogenized, and added with 60% aqueous ethanol solution at liquid: solid ratio of 1:5 (v/w). After 1h of continuous stirring, the extraction process was carried out in closed bottle at room temperature (20-22 °C), in dark conditions, for 30 days. The hazelnut suspension was mixed by hand each three days. At the end of the maceration procedure, the hazelnut liquid extract (HZN) was collected, passed through a filter of 0.2μm and stored at -80°C for all experiments. The complete metabolomic characterization of the HZN ethanolic liquid extract used in this study was performed by liquid chromatography-high resolution mass spectrometry [14].
Cell cultures and treatments
HepG2 human hepatocellular carcinoma cells (RRID: CVCL_0027) were purchased from the European Collection of Cell Cultures (ECACC, Sigma-Aldrich), maintained as subconfluent monolayers in DMEM, with 10% heat-inactivated foetal bovine serum, 2mM glutamine, 1% non-essential amino acids and 1% penicillin-streptomycin (10,000U/mL), at 37°C in a 5% CO2 atmosphere in air. THLE-2 cells (RRID: CVCL_3803), derived from SV40-immortalized normal human liver cells, were purchased from the American Type Culture Collection, ATCC (Manassas, VA, USA). They were cultured in LHC-9 medium, supplemented with 70ng/mL phosphoethanolamine, 5ng/mL epidermal growth factor, 10% fetal bovine serum (Hyclone) and antibiotics as above. Flasks and dishes for THLE-2 cultures and experiments were pre-coated with collagen (2.9mg/mL), fibronectin (1mg/mL) and bovine serum albumin (1mg/ mL) according to ATCC guidelines. In all the experiments, cells were seeded in 35mm Petri-dishes at 2x105 cells/dish; 24h after plating, cells were treated with either the Hazelnut (HZN) ethanolic extract (ranging from 0.004 to 0.4 mg/ml) or the corresponding aqueous ethanol solution (0.0006-0.6 %) for 72h (without any medium replacement) (Figure 1a). The solutions were freshly prepared before each experiment in culture media. The effect on cell proliferation was assessed by counting viable cells at the hemocytometer following Trypan blue staining (0.4% solution). In the experiments with the DAC, the agent was dissolved in DMSO, freshly diluted in PBS before each experiment, and added to cells at final concentration of 0.5μM for 24 h (a non-toxic concentration for THLE-2 cells, data not shown).
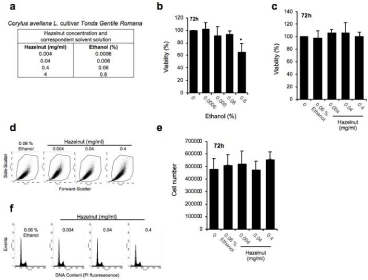
Figure 1: The Corylus avellana L. extract is not toxic to THLE-2 human primary hepatocytes. (a) Table summarizing the equivalence between the different hazelnut
concentrations (mg/ml) and the corresponding ethanol solution (%) used in the experiments. (b) Evaluation of cell viability percentage, carried out in THLE-2 cells
by haemocytometer counting of trypan blue negative cells, following treatment with the reported ethanol concentrations (administered for 72h). *P<0.05 in 0.6%
ethanol compared with the untreated control, N=3 independent experiments. (c) Cell viability assessment (trypan blue counting), and (d) flow cytometric dot plots
showing the size/morphology (estimated by FSC-H versus SSC-H parameters) of THLE-2 cells undergoing either control ethanol (0.06% Eth) and hazelnuts
treatments (expressed in mg/ml). Gated cells represent integer cells. (e) Analysis of cell proliferation carried out by haemocytometer counting of trypan blue
negative cells. (f) FACS (fluorescence-activated single cell sorti) analysis of cell cycle distribution in ethanol-fixed cells stained with Propidium Iodide (PI).
Flow cytometric analysis
A FACScan flow cytometer (Becton Dickinson, Bedford, MA, USA) equipped with a 488nm argon laser, was used for all flowcytometric analyses. Forward (FSC-H) and side scatters (SSC-H) parameters were used to establish cell size and granularity parameters, respectively, and to delimitate the integer/healthy cells (identified by the gate) and exclude cellular debris.
The analysis of the DNA content for cell cycle distribution was carried out by applying the FlowJo software® as previously described [4], by staining the 80% (v/v) ethanol-fixed cells with a mix containing propidium iodide (PI, 50μg/ml), RNAse A (50μg/ml), Triton X-100 (0.1%), EDTA (0.1mM) in PBS.
The evaluation of the LDLR membrane protein expression by fluorescent antibody staining (Alexa-Fluor® 488-conjugated monoclonal antibody and the isotype control antibody, R&D systems, Minneapolis, USA) was carried as previously reported [4]. Briefly, adherent cells were harvested by cold trypsinization, washed twice in a solution containing ice cold PBS, 10% foetal calf serum, and 1% sodium azide (PSSA solution), and adjusted to a final concentration of 1×106 cells/ml for staining with the antibody. Either the human LDLR Alexa-Fluor® 488-conjugated monoclonal antibody or the isotype control antibody (R&D systems) were added to cell solution at the concentration of 5μl/106 cells (2 h staining in the dark, at 4°C). For fluorescence data analysis, the Mean Fluorescence Intensity (MFI) was calculated as the delta between the antibody-labelled cells and the unstained ones.
RNA extraction, reverse transcription and gene expression analysis
Total RNA was extracted by Trizol® (Invitrogen, Thermo Fisher Scientific) followed by spin-column elution, also including a DNAse I digestion step (Direct-zolTM RNA miniPrep, Zymoresearch, Irvine, CA, USA), as previously described [4]. TaqMan® Reverse Transcription Reagent (Applied Biosystems, Thermo Fisher Scientific) was used to perform retro-transcription of total RNA with random primers. Analysis of the LDLR expression was carried out as previously described [4], by normalizing to the Glyceraldehyde- 3-Phosphate Dehydrogenase (GAPDH) expression, applying the 2-ΔΔCt method [15]. PCR primers were designed by NCBI-Primer Blast free software (https://www.ncbi.nlm.nih.gov/tools/primerblast), according to gene sequences available in the UCSC database (https://genome.ucsc.edu), and selected to amplify an exon-intronexon region (≤200bp) to exclude genomic contamination. PCR primers were synthesized by Eurofins Scientific (Luxembourg). The primer sequences are the following: hsa LDLR (RefSeq NM_000527); forward (5’-3’): TCAGTTCTGGAGGTGCGATG; reverse (5’- 3’): TGAGTCACAGACGAACTGCC; hsa GAPDH (RefSeq NM_002046); forward (5’-3’): GCACCGTCAAGGCTGAGAAC; reverse (5’-3’): GAGGGATCTCGCTCCTGGA.
DNA extraction and DNA methylation analysis of LDLR gene promoter
DNA methylation analysis was performed by bisulfite conversion and pyrosequencing, as previously detailed [4,16]. Genomic DNA was extracted from liver cells using the Quick-DNATM Miniprep kit (Zymo Research, Irvine, CA, USA). Five hundred nanograms underwent bisulfite modification using the EZ DNA Methylation- GoldTM Kit (Zymo Research) according to the manufacturer’s protocol. Bisulfite-converted DNA (50ng in 50μl reaction) was amplified using the One Taq® Quick-Load® 2×Master Mix (New England Biolabs, Ipswich, MA, USA), and pyrosequencing was performed using the Pyromark® Q24 Instrument (Qiagen, Hilden, Germany). The percentage of methylation, expressed as percentage of 5-methyl cytosine (5-mC), was calculated by the provided Qiagen software, and expressed as the number of 5-mC divided by the sum of methylated and unmethylated cytosines. One investigator, blind to all the information regarding the samples except their code, performed the pyrosequencing analyses. Different internal controls were included in every pyrosequencing run to ensure the completion of bisulfite modification, the specificity of PCR amplification, and the success of pyrosequencing reactions. A universal methylated DNA standard (Zymo Research) was used as a positive control for bisulfite conversion.
To study the DNA methylation of the human LDLR promoter, we selected two previously identified regions [4]: one mapping at position -221/+174 with respect to the transcription starting site (TSS) (fragment #1), containing nine cytosines (identified by Y1-Y9); a second one mapping at position -739/-548 on the LDLR promoter gene (fragment #2), with four cytosines (Y1-Y4).
Statistical analysis
The variation of samples values is reported as Mean ± S.D. (standard deviation), calculated in N=3 independent experiments. The statistical differences were analyzed trough the KailedaGraph software (Synergy Software, Reading PA, USA) by applying the one-way analysis of variance (ANOVA) followed by Tukey’s test for multiple comparisons. P values <0.05 were considered statistically significant and indicated as follows: *P <0.05; **P <0.01.
Results
The Corylus avellana L. ethanolic extract is not toxic to THLE-2 human primary hepatocytes
We treated the THLE-2 cells with increasing concentrations of the ethanolic extract of C. avellane (HZN) to first evaluate the impact on cell integrity, viability and proliferation. Given the well-documented sensitivity of liver cells to ethanol (here used as solvent to prepare the hazelnut solution in the range of 0.0006-0.6% [4,14]) (Figure 1a), we preliminarily assessed the viability of THLE-2 cells exposed to ethanol for 72 hours. As reported in (Figure 1b), the highest ethanol concentration (0.6%) reduced cell viability by ~60% and was thus excluded (along with the correspondent 4mg/ml of HZN) in the following experiments. Moreover, (Figure 1c-1d), no change in cell viability (Figure 1c) and cell integrity (as assessed by fluorescenceactivated single cell sorting analysis, FACS) (Figure 1d) was reported in cells undergoing 72 h treatment with HZN up to 0.4mg/ml. Accordingly, no significant modulation of cell proliferation (Figure 1e) and cell cycle distribution (Figure 1f) was observed in HZNtreated cells if compared to ethanol solvent solution.
The LDLR expression and DNA promoter methylation of THLE-2 human primary hepatocytes significantly differ from HepG2 hepatocarcinoma cell line
To verify whether any difference in terms of LDLR gene expression occurs in THLE-2 cells compared to HepG2, we measured LDLR transcript level by real-time PCR, and demonstrated that LDLR expression is lower in primary hepatocytes if compared to carcinoma cells at basal level (Figure 2a). By treating THLE-2 cells with the specific demethylating agent 5-Aza-2-deoxycytidine (DAC, 0.5μM for 24 h), we further prove that the different LDLR basal level between cell lines is due to a dissimilar DNA promoter methylation pattern, as the forced hypo-methylation by DAC significantly increases the LDLR mRNA expression in THLE-2 cells (Figure 2a). To confirm this finding, we carried out DNA methylation analysis of two regions in the human LDLR gene promoter (Figure 2b), rich in multiple cytosine sites (indicated with Y) [detailed in ref. 4]. As reported in (Figure 2c), fragment #1 displays a very low degree of methylation in HepG2, consistent to what previously reported [4], the average percentage of cytosine methylation (% 5-mC) being 5.4±1.6. The methylation pattern of fragment #1 in primary THLE-2 (25.4±1.5 of % 5-mC) is much higher than what reported in HepG2, showing a massive DNA methylation percentage in all cytosine positions (Figure 2c). Such difference between cell lines is even higher in fragment #2, the LDLR promoter being strongly hyper-methylated in THLE-2 cells (over 57% of 5-mC) if compared to HepG2 pattern (7.5±1.2 % 5-mC) (Figure 2d).
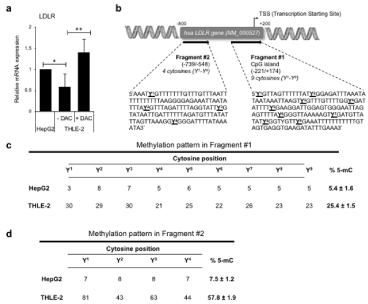
Figure 2: The LDLR expression and DNA promoter methylation of THLE-2 human primary hepatocytes significantly differ from HepG2 hepatocarcinoma cell line.
(a) Evaluation of low-density lipoprotein receptor (LDLR) gene expression carried out by real-time PCR in HepG2 and THLE-2 cells, 72 h after plating. THLE-2 cells
have been also treated with the 5-Aza-2-Deoxycytidine (0.5μM DAC), 24 h before expression assessment. (b). Schematic representation of the human LDLR gene
promoter, where the two regions selected for DNA methylation analysis (fragment #1 and #2), are indicated. The putative cytosine methylation sites have been
identified by YN, with N indicating the progressive position number along gene promoter. CpG, cytosine-guanine dinucleotide separated by a phosphodiester bond;
hsa, homo sapiens; TSS: Transcription Starting Site. (c,d) DNA methylation pattern of fragment #1 and #2 obtained by bisulfite conversion and pyrosequcning. The
methylation percentage for each cytosine site is reported. The % 5-mC (5-methyl cytosine, right column in the table) represents the average methylation percentage
of the whole fragment, and it is expressed as mea Standard Deviation (SD) in N=3 independent experiments.
The ethanolic extract of C. avellana stimulates LDLR expression by hypo-methylating both #1 and #2 promoter regions in THLE-2 cells
To characterize the epigenetic response of primary hepatocytes to hazelnut administration, LDLR expression and promoter DNA methylation were evaluated in response to HZN in primary hepatocytes. As shown in (Figure 3a), all HZN doses (0.004-0.4 mg/ ml, administered for 72 h) trigger a significant stimulation of LDLR gene expression if compared to control treatment (ethanol 0.06%). This increment is consistent with the stimulation of LDLR membrane protein expression in the HZN-compared to ethanol-treated cells, as assessed by staining cells with a specific LDLR-Alexa-Fluor® 488-conjugated antibody (Figure 3b). In terms of DNA methylation level, we prove a significant decline of the average % 5-mC in cells undergoing HZN administrations (72h treatment) at both fragment #1 and #2 level (Figure 3c). In details, the pyrosequencing data (Figure 3d) show that cytosines Y2, Y4, Y5 and Y7 are the most sensitive ones to the demethylating function triggered by the hazelnut extract in the fragment closest to the TSS. In fragment #2 the hypo-methylation mostly affects cytosine Y2, whereas other sites strongly retain their methylation pattern (Figure 3e).
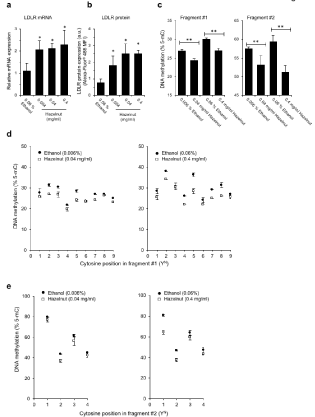
Figure 3: The ethanolic extract of C. avellana stimulates LDLR expression by hypo-methylating both #1 and #2 promoter regions in THLE-2 cells. Evaluation of the
Low-Density Lipoprotein Receptor (LDLR) expression, in terms of (a) transcript level and (b) membrane protein level (a.u., arbitrary units), carried out in THLE-2
cells following 72 h treatment with the indicated hazelnut extract doses (HZN, mg/ml) and ethanol (Eth) control administration. *P<0.05 in hazelnut- versus ethanoltreated
samples, N=3 independent experiments. (c) Assessment of the DNA methylation pattern (% 5-mC, 5-methyl cytosine) carried out in fragment #1 and #2
of the LDLR gene promoter in THLE-2 cells following 72 h treatment with either hazelnut or ethanol control solution. **P<0.01 in hazelnut- versus ethanol-treated
samples, N=3 independent experiments. (d,e) DNA methylation pattern of each single cytosine position mapping in fragment #1 and fragment #2 of human LDLR
gene promoter.
Discussion
We here demonstrate that LDLR expression is epigenetically regulated in human liver by DNA methylation, and that the epigenetic pattern is differently regulated by gene promoter methylation in THLE-2 primary liver cells if compared to HepG2 hepatocarcinoma. Moreover, in response to hazelnut (C. avellana L), primary liver cells stimulate LDLR expression via a strong DNA hypo-methylation that involves two regions at promoter levels, further supporting the occurrence of a hazelnut-specific epigenetic signature that is much stronger -and physiologically more relevant-in primary hepatocytes if compared to HepG2 cancer cells. In liver, the LDLR triggers the endocytosis of the LDL particles from plasma, thus regulating the blood cholesterol levels [17]. By characterizing such positive regulation by hazelnut on hepatic LDLR expression, our experimental findings provide new clues on the molecular pathways underlying the health benefits exerted by hazelnut consumption on lipid content and liver metabolism [1,18].
We here directly compared HepG2 carcinoma and THLE-2 primary liver cells in terms of LDLR basal expression and DNA methylation promoter pattern, with the objective to better characterize the ability of a hazelnut extract to modulate LDLR expression in liver by using a physiological experimental model. In a recently paper by our group [4], we identified the first epigenetic pathway triggered by nut species in HepG2 liver cells. We demonstrated that LDLR expression is stimulated by C. avellana, and that this increase is in part due to a small but significant hypomethylation in a TSS-distant region of the LDLR gene promoter. Multiple studies have shown that the level of DNA methylation in promoters correlates with the expression of downstream genes, the DNA hypomethylation of gene promoters being associated to the stimulation of transcription [19]. Our experimental findings shed the light on the possible epigenomics involvement of hazelnuts consumption that might help disclosing new functional properties of nuts. Our previous data in HepG2 cells needed to be validated and corroborated in a more physiological experimental model, to exclude the hypothesis it might be limited to hepatocarcinoma cells. We indeed demonstrated that LDLR is differently regulated in HepG2 versus THLE-2, the former displaying a higher basal expression level. Reduced mRNA content in primary liver cells is due to a strong epigenetic lock that DNA methylation exerts on LDLR gene promoter. Both TSS-near (fragment #1) and TSS-far (fragment #2) regions display a significant reduced methylation pattern in HepG2 compared to THLE-2, the former cells resembling what reported in housekeeping genes. The percentage of methylated cytosines in HepG2 is very low, especially in the regions closed to the TSS, to allow the transcription machine to access the promoter.
These findings are line with the well-documented epigenome rearrangement occurring in transformed versus primary cells that has been extensively demonstrated in different cancer histotypes, including liver [20-22]. Other authors reported epigenetic divergences in HepG2 compared with THLE cells, in terms of microRNA and chromatin changes [23,24]. As here demonstrated in LDLR gene promoter, these two cells lines also differ at DNA methylation level. Future high-throughput epigenetic analyses might help providing a more extensive description of DNA methylation differences that need to be considered in the selection of the proper experimental model for studies of liver metabolism. Epigenetics tunes liver response to drugs and metabolites, characterizes pathological conditions [25,26], and might hence differently affect the biological output according to the in vitro cellular context.
Food components have been also proven to affect liver physiology by epigenetic changes [27,28]. In order to set-up new studies addressing the impact of nutrition on liver cells, the epigenetic characterization of hepatic cell lines, such as HepG2 and THLE-2, need hence to be defined. We here characterized the effect driven by an ethanolic extract of C. avellana on the DNA methylation pattern of the LDLR gene, and demonstrated that the epigenetic response to nuts is significantly amplified in THLE-2 cells compared to hepatocarcinoma. The epigenetic property exerted by hazelnut is much stronger in primary cells than HepG2; nuts administration is responsible for a strong hypomethylation of both fragment #1 and #2 regulatory regions in primary liver cells, whereas in HepG2 we reported an epigenetic effect exclusively in the TSS-distant region [4]. We previously commented that LDLR stimulation in HepG2 should be explained in terms of additional mechanisms, still to be identified. In primary liver cells, hazelnut treatment impacts on both TSS-closed and TSS-far regulatory region, demonstrating the occurrence of a global epigenetic effect on the whole promoter region of LDLR gene.
The average promoter methylation is usually adopted in studies to quantify and compare the DNA methylation pattern. Under our experimental conditions, we indeed demonstrated that % 5-mC significantly differ between HepG2 and THLE-2 at basal level, as well as in response to hazelnut. However, we identified and mapped some cytosines that seem to be more sensitive to the hazelnut-induced effect. Recent studies suggested that methylation of individual cytosines can be determinant to drive transcription [29]. It was also shown that a single differentially methylated CpG might affect transcription of the ESR1 gene [30]. Future possible investigations by site-specific mutation analysis might help verifying whether this is the case also in LDRL gene promoter of liver cells.
Conclusion
Overall, our data demonstrate that the epigenetic regulation controlling LDLR gene promoter via DNA methylation- differ in THLE-2 primary liver cells compared with hepatocarcinoma, and that the ethanolic extract of Corylus avellana L. stimulates LDLR expression through the hypomethylation of two regulatory regions in primary hepatocytes.
As the involvement of DNA methylation changes by food components in liver cells is gaining interest in terms of impact on human health and prevention to pathologies, our experimental findings might put the bases for further characterization of the epigenetic differences occurring in different in vitro liver experimental models, that need to be considered in future studies for reliable assessment of nutrition effect on liver physiology and metabolism.
References
- Chen GC, Wan ZX, Qin LQ. Nut consumption, lipid profile, and health outcomes. Am J Clin Nutr. 2016; 103: 1185-1186.
- Berryman CE, Fleming JA, Kris-Etherton PM. Inclusion of Almonds in a Cholesterol-Lowering Diet Improves Plasma HDL Subspecies and Cholesterol Efflux to Serum in Normal-Weight Individuals with Elevated LDL Cholesterol. J Nutr. 2017; 147: 1517-1523.
- Santi C, Giorni A, Terenzi CT, Altavista P, Bacchetta L. Daily hazelnut intake exerts multiple reversible effects on plasma profile of healthy subjects. Food and Nutrition Sciences. 2017; 8: 633-646.
- Benassi B, Santi C, Santangeli S, Grollino MG, Raschellà G, Bacchetta L, et al. Modulation of LDL receptor expression and promoter methylation in HepG2 cells treated with a Corylus avellana L. J Funct Foods. 2019; 53: 208-218.
- Nikoloff N, Larramendy ML, Soloneski S. Assessment of DNA damage, cytotoxicity, and apoptosis in human hepatoma (HepG2) cells after flurochloridone herbicide exposure. Food Chem Toxicol. 2014; 65: 233e241.
- Kaur P, Robin, Mehta RG, Arora S, Singh B. Progression of conventional hepatic cell culture models to bioengineered HepG2 cells for evaluation of herbal bioactivities. Biotechnol Lett. 2018; 40: 881-893.
- Hewitt NJ, Lechón MJ, Houston JB, Hallifax D, Brown HS, Maurel P, et al. Primary hepatocytes: current understanding of the regulation of metabolic enzymes and transporter proteins, and pharmaceutical practice for the use of hepatocytes in metabolism, enzyme induction, transporter, clearance, and hepatotoxicity studies. Drug Metab Rev. 2007; 39: 159-234.
- Soltanpour Y, Hilgendorf C, Ahlstrom MM, Foster AJ, Kenna JG, Petersen A, et al. Characterization of THLE-Cytochrome P450 (P450) Cell Lines: Gene Expression Background and Relationship to P450-Enzyme Activity. Drug Metab Dispos. 2012; 40: 2054-2058.
- Pfeifer AM, Cole KE, Smoot DT, Weston A, Groopman JD, Shields PG, et al. Simian virus 40 large tumor antigen-immortalized normal human liver epithelial cells express hepatocyte characteristics and metabolize chemical carcinogens. Proc Natl Acad Sci USA. 1993; 90: 5123-5127.
- Tran DDH, Koch A, Allister A, Saran S, Ewald F, Koch M, et al. Treatment with MAPKAP2 (MK2) inhibitor and DNA methylation inhibitor, 5-aza dC, synergistically triggers apoptosis in hepatocellular carcinoma (HCC) via tristetraprolin (TTP). Cell Signal. 2016; 28: 1872-1880.
- Buurman R, Gürlevik E, Schäffer V, Eilers M, Sandbothe M, Kreipe H, et al. Histone deacetylases activate hepatocyte growth factor signaling by repressing microRNA-449 in hepatocellular carcinoma cells. Gastroenterology. 2012; 143: 811-820.
- Alvarez ML, Khosroheidari M, Eddy E, Done SC. MicroRNA-27a decreases the level and efficiency of the LDL receptor and contributes to the dysregulation of cholesterol homeostasis. Atherosclerosis. 2015; 242: 595-604.
- Cai D, Yuan M, Liu H, Pan S, Ma W, Hong J, et al. Maternal betaine supplementation throughout gestation and lactation modifies hepatic cholesterol metabolic genes in weaning piglets via AMPK/LXR-mediated pathway and histone modification. Nutrients. 2016; 8: 646.
- Cappelli G, Giovannini D, Basso A, Demurtas OC, Diretto G, Santi C, et al. A Corylus avellana L. extract enhances human macrophage bactericidal response against Staphylococcus aureus by increasing the expression of anti-in- flammatory and iron metabolism genes. J Funct Foods. 2018; 45: 499-511.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001; 25: 402- 408.
- Consales C, Cirotti C, Filomeni G, Panatta M, Butera A, Merla C, et al. Fifty- Hertz Magnetic Field Affects the Epigenetic Modulation of the miR-34b/c in Neuronal Cells. Mol Neurobiol. 2018; 55: 5698-5714.
- Goldstein JL, Brown MS. History of Discovery: The LDL receptor. Arterioscler Thromb Vasc Biol. 2009; 29: 431-438.
- Sabate, J, Ros E, Salas-Salvado J. Nuts: Nutrition and health outcomes. Br J Nutr. 2006; 96: S1.
- Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001; 293: 1068-1070.
- Rakyan VK, Down TA, Balding DJ, Beck S. Epigenome-wide asso-ciation studies for common human diseases. Nature Reviews Genetics. 2011; 12: 529.
- Shen J, Wang S, Zhang YJ, Kappil M, Wu HC, Kibriya MG, et al. Genomewide DNA methylation profiles in hepatocellular carcinoma. Hepatology. 2012; 55: 1799-1808.
- Toh TB, Lim JJ, Chow EK. Epigenetics of hepatocellular carcinoma. Clin Transl Med. 2019; 8: 13.
- Kirchmeyer M, Servais FA, Hamdorf M, Nazarov PV, Ginolhac A, Halder R, et al. Cytokine-mediated modulation of the hepatic miRNome: miR-146b-5p is an IL-6-inducible miRNA with multiple targets. J Leukoc Biol. 2018; 104: 987-1002.
- Buurman R, Gürlevik E, Schäffer V, Eilers M, Sandbothe M, Kreipe H, et al. Histone deacetylases activate hepatocyte growth factor signaling by repressing microRNA-449 in hepatocellular carcinoma cells. Gastroenterology. 2012; 143: 811-820.
- Lee SM, Kim-Ha J, Choi WY et al. Interplay of genetic and epigenetic alterations in hepatocellular carcinoma. Epigenomics. 2016; 8: 993-1005.
- Ling C, Rönn T. Epigenetics in Human Obesity and Type 2 Diabetes. Cell Metab. 2019; 29: 1028-1044.
- Moreno FS, Heidor R, Pogribny IP. Nutritional Epigenetics and the Prevention of Hepatocellular Carcinoma with Bioactive Food Constituents. Nutr Cancer. 2016; 68: 719-733.
- Kim H, Worsley O, Yang E, Purbojati RW, Liang AL, Tan W, et al. Persistent changes in liver methylation and microbiome composition following reversal of diet-induced non-alcoholic-fatty liver disease. Cell Mol Life Sci. 2019; 76: 4341-4354.
- Medvedeva YA, Khamis AM, Kulakovskiy IV, Ba-Alawi W, Bhuyan MS, Kawaji H, et al; FANTOM consortium. Effects of cytosine methylation on transcription factor binding sites. BMC Genomics. 2014; 15: 119.
- Furst RW, Kliem H, Meyer HH, Ulbrich SE. A differentially methylated single CpG-site is correlated with estrogen receptor alpha transcription. J Steroid Biochem Mol Biol. 2012; 130: 96-104.