Clinical Image
Austin J Nutri Food Sci. 2018; 6(1): 1096.
Micrornas for Molecular Screening of Colon Cancer Early Stages, Following Mild Food Intakes: Statistical and Bioinformatical Considerations
Ahmed FE¹*, Vos PW², Ahmed NC¹, Gouda M³ and Hussein L³
¹GEM Tox Labs, Institute for Research in Biotechnology, USA
²Department of Biostatistics, College of Allied Health Sciences, East Carolina University, USA
³Department of Nutrition & Food Science, National Research Center, Egypt
*Corresponding author: Farid E. Ahmed, GEM Tox Labs, Institute for Research in Biotechnology, 2905 South Memorial Drive, Greenville, NC 27834, USA
Received: January 05, 2018; Accepted: February 08, 2017; Published: February 19, 2018
Clinical Image
Colon Cancer (CC) is curable at the early disease stages (0-2), and therefore early screening is essential for good outcome, and reduces patients’ mortality. Early detection would be desirable, if accurate and cost effective diagnostic procedures are available, and would result in better patient outcomes.
Today, the often employed immunological fecal occult blood test (FOBTi) does not have adequate sensitivity and necessitates food restriction, resulting in a decreased compliance. Colonoscopy, a reliable method to screen for that cancer, and which is considered as the “Gold Standard” for CC screening is invasive, often accompanied by abdominal pain, has potential for complications and is not economical, and in certain cases it could lead to mortality, all are negative factors that have hampered its application worldwide.
On the other hand, a screening approach that utilizes the relatively stable and nondegradable micro RNA molecules --when extracted from either the noninvasive human stool, or the semiinvasive blood samples by available commercial kits and manipulated thereafter-- would be more preferable diagnostic screening test than a transcriptomic messenger (m) RNA-, a mutation DNA-, an epigeneticor a proteomic-based test. That approach utilizes reverse transcriptase (RT), followed by a modified quantitative real-time polymerase chain reaction (qPCR). Because exosomal miRNAs will not be determine, an additional test needs to be carried out simultaneously on stool or plasma’s total small RNAs, and subsequently corrections are carried out to compensate for the undetermined exosomes, in order to obtain valid test results. Ultimately, a chip would be developed to facilitate diagnosis, as has been carried out for the quantification of Genetically Modified Organisms (GMOs) in foods. If the performance criteria carried out by laboratories to enure proper complaisance are not achieved, then quantifying miRNA in stool or blood samples using high throughput automated technologies --similar to measurements used in the clinical laboratory-would eventually be advanced to the clinical setting, making a noticeable impact on the prevention of colon cancer.
There are many advantages of testing human stool compared to other colon cancer testing methods because of its noninvasive nature, and lack of requirement to cathartic methods, which are often unpleasant, no visits to health care personnel to take testing material because a person can collect his or her stool at their homes, or taking time off from work and any other activities. Unlike sigmoidoscopy, it reflects the full length of the colorectum and samples can be taken in a way that represents both the right, as well as the left side of the colon. Because colonocytes are continuously and abundantly ejected from gut lumen into the fecal stream, as opposed to what occurs in blood --where the release is intermittently-as is the situation FOBT, and transformed colonoctes produce more RNA than normal ones; therefore, this natural enrichment phenomenon partially obviate for the need to use a laboratory technique to enrich for tumorigenic colonocytes. Moreover, because specimens can be mailed to Laboratories for testing, this results in an unimpeded geographic access to the stool for screening purposes.
It should be emphasized that although shed cells in stool are derived from tumor and also from ordinary tissue, current research data indicate that adequate circulating. miRNA genes (either cellular or extra cellular) diagnostic miRNA gene expression profiles are correlated with enough number of exfoliated cancerous cells. Moreover an abundance of transformed RNA is released in the stool, and can also be measured circulating in blood, which can be determined quantitatively by a sensitive technique such as PCR in spite of the presence of bacterial DNA, non-transformed RNA and other interfering substances. MiRNA quantification is feasible due to the high specificity of employed PCR primers, overcoming these obstacles; therefore, the number of abnormally-shed colonocytes in stool, or total RNA presents in plasma or serum becomes unlimiting.
A test that employs miRNAs in stool or blood could also result in a robust screen because of the durability of the miRNA molecules. Moreover, an approach utilizing miRNA genes is more comprehensive and encompassing than a test that is based on the fragile messenger (m) RNA, for example, because it is based on mechanisms at a higher level of control. We believe that ultimately the final noninvasive test in stool or blood will include testing of several miRNA genes that show increased and decreased expression, and eventually a chip that contains a combination of these stable molecules will be produced to simplify testing.
Commercial preparations are available for testing of mature miRNAs, which saves valuable time, as well as allow use robust manufacturer’s established validation and QC standards. Small noncoding RNAs that exhibit little variation in different cell types (e.g., snoRNAs and snRNAs) are polyadenylated and are reverse transcribed (RT) in the same way as the small miRNAs and thereby could serve as controls for variability in sample loading and realtime RT-PCR efficiency. They are, however, not suited for data normalization in miRNA profiling experiments because they are not well expressed in serum and plasma samples. Therefore, normalization by a plate mean (i.e., mean CT value of all the miRNA targets on the plate), or using a commonly expressed miRNA targets (i.e., only the targets that are expressed in all samples are used to calculate the mean value) would be needed for a proper normalization of the amplification reaction. As sensitive and quantitative testing methods such as NGS, microarrays- and qPCR-based approaches are now available; colon cancer-specific miRNAs are identified in stool colonocytes or blood plasma by informatics’ validation of miRNA/mRNA target pairs within pathways of interest could lead to discovery of cellular functions collectively targeted by differentially expressed miRNAs.
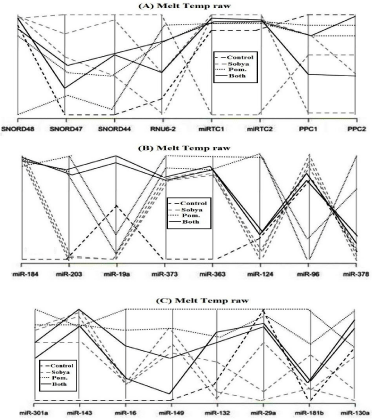
Contrary to screening of large numbers of mRNA genes, a small number of miRNAs is used to differentiate between cancer and normal conditions, and unlike mRNA, miRNAs in stool and blood remain largely intact and stable for detection. Therefore, miRNAs are better molecules to use for developing a reliable noninvasive diagnostic screen for colon cancer, since we found out that: a) the presence of Escherichia coli does not hinder detection of miRNA by a sensitive technique such as qPCR, as the primers employed are selected to amplify human and not bacterial miRNA genes, and b) the miRNA expression patterns are the same in primary tumor, or in diseased tissue, as in stool and blood samples. Although exosomal RNA will be missed when using restricted extraction of total RNA from blood or stool, a parallel test could also be carried out on the small total RNA of the seven miRNAs (miR-184, miR-203, miR- 373, miR-124, miR-96, miR-373 and miR-301a), which separated candidate miRNAs, because of different separation melting profiles. These miRNAs could thus function as novel molecular markers of relevance to oxidative stress and immunoglobulin function, for the intake of polyphenol (PP)-rich, functional fermented foods rich in lactobacilli (FS), or their combination (Figure 1), which is a graphical representation of the parallel plot ccoordinates of the studies miRNA genes. The genes were ordered using the p-values of a one way ANOVA based on groups. Genes with the smallest p-values are presented first. Figure 1A below shows control genes, while in Figures 1B, and C, five miRNA genes show separation.